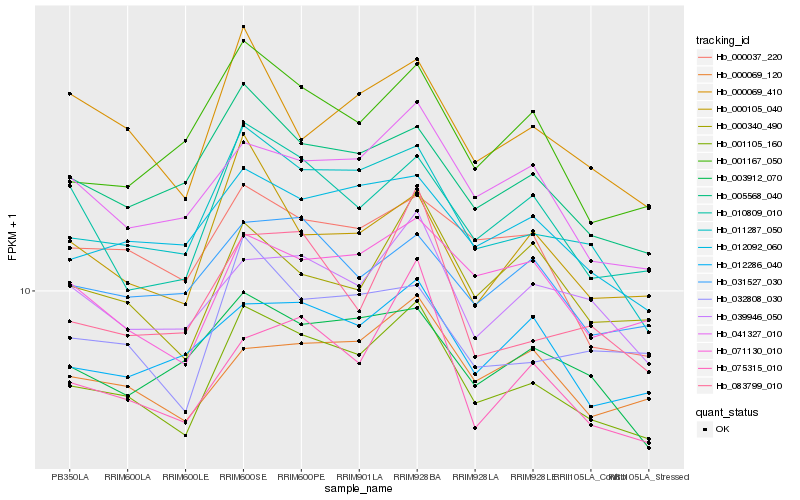
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001105_160 |
0.0 |
- |
- |
PREDICTED: protein phosphatase 2C 32 [Jatropha curcas] |
| 2 |
Hb_011287_050 |
0.057380786 |
- |
- |
PREDICTED: uncharacterized protein LOC105644786 [Jatropha curcas] |
| 3 |
Hb_041327_010 |
0.0770322491 |
- |
- |
PREDICTED: la-related protein 1A isoform X1 [Jatropha curcas] |
| 4 |
Hb_005568_040 |
0.0778212439 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X2 [Jatropha curcas] |
| 5 |
Hb_012092_060 |
0.0790851293 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 [Jatropha curcas] |
| 6 |
Hb_071130_010 |
0.0803396003 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL1 isoform X1 [Jatropha curcas] |
| 7 |
Hb_010809_010 |
0.080583292 |
- |
- |
Na+/H+ antiporter [Populus euphratica] |
| 8 |
Hb_039946_050 |
0.0817589903 |
- |
- |
catalytic, putative [Ricinus communis] |
| 9 |
Hb_032808_030 |
0.0824615258 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_001167_050 |
0.0853734182 |
- |
- |
PREDICTED: uncharacterized protein LOC105648014 [Jatropha curcas] |
| 11 |
Hb_012286_040 |
0.0854288268 |
- |
- |
PREDICTED: pre-mRNA-splicing factor RSE1 [Jatropha curcas] |
| 12 |
Hb_000069_120 |
0.0857189527 |
- |
- |
PREDICTED: uncharacterized protein LOC105124742 isoform X1 [Populus euphratica] |
| 13 |
Hb_000340_490 |
0.0862952134 |
- |
- |
PREDICTED: ankyrin-1-like [Jatropha curcas] |
| 14 |
Hb_000105_040 |
0.0872770859 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOR isoform X2 [Jatropha curcas] |
| 15 |
Hb_003912_070 |
0.0873786175 |
- |
- |
PREDICTED: nuclear pore complex protein NUP85 [Jatropha curcas] |
| 16 |
Hb_000069_410 |
0.0880355336 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 17 |
Hb_031527_030 |
0.0883526559 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |
| 18 |
Hb_000037_220 |
0.0883698353 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 11 homolog [Jatropha curcas] |
| 19 |
Hb_075315_010 |
0.0886019861 |
- |
- |
ornithine aminotransferase [Camellia sinensis] |
| 20 |
Hb_083799_010 |
0.0893611934 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |