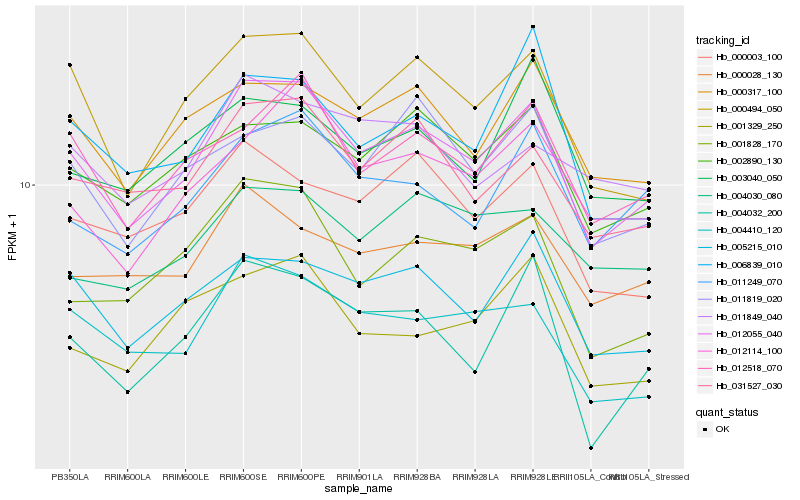
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012055_040 |
0.0 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 2 |
Hb_000494_050 |
0.0572456281 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 3 |
Hb_012518_070 |
0.0603335521 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 9 isoform X1 [Jatropha curcas] |
| 4 |
Hb_031527_030 |
0.0615932931 |
- |
- |
PREDICTED: ribosomal RNA processing protein 1 homolog [Jatropha curcas] |
| 5 |
Hb_005215_010 |
0.0651427615 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 6 |
Hb_004410_120 |
0.0657208209 |
- |
- |
PREDICTED: uncharacterized protein LOC105633877 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000317_100 |
0.0670423347 |
- |
- |
PREDICTED: uncharacterized protein LOC105646995 [Jatropha curcas] |
| 8 |
Hb_002890_130 |
0.0673193164 |
- |
- |
tip120, putative [Ricinus communis] |
| 9 |
Hb_011249_070 |
0.0680627472 |
- |
- |
hypothetical protein POPTR_0018s12610g [Populus trichocarpa] |
| 10 |
Hb_012114_100 |
0.0686144935 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 11 |
Hb_011819_020 |
0.0691512244 |
- |
- |
PREDICTED: uncharacterized protein LOC105643703 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001828_170 |
0.0701037741 |
- |
- |
PREDICTED: protein TPLATE [Jatropha curcas] |
| 13 |
Hb_004032_200 |
0.0721761112 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105635137 [Jatropha curcas] |
| 14 |
Hb_000028_130 |
0.0727053142 |
- |
- |
PREDICTED: uncharacterized protein LOC105645303 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000003_100 |
0.0748732965 |
transcription factor |
TF Family: SNF2 |
PREDICTED: transcription termination factor 2 isoform X5 [Jatropha curcas] |
| 16 |
Hb_006839_010 |
0.0759114313 |
- |
- |
PREDICTED: importin subunit beta-1-like [Jatropha curcas] |
| 17 |
Hb_001329_250 |
0.0774708793 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105642692 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004030_080 |
0.079183513 |
- |
- |
hypothetical protein JCGZ_25110 [Jatropha curcas] |
| 19 |
Hb_011849_040 |
0.0800639309 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 20 |
Hb_003040_050 |
0.0803415669 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |