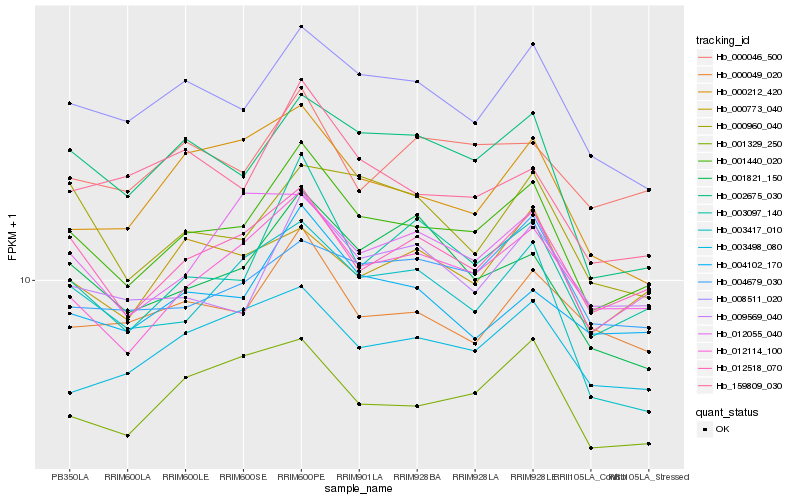
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001440_020 |
0.0 |
- |
- |
exocyst componenet sec8, putative [Ricinus communis] |
| 2 |
Hb_012518_070 |
0.0597552391 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 9 isoform X1 [Jatropha curcas] |
| 3 |
Hb_012114_100 |
0.063073553 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 4 |
Hb_008511_020 |
0.0639542997 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 5 |
Hb_003097_140 |
0.06822887 |
- |
- |
PREDICTED: glycosyltransferase-like KOBITO 1 [Jatropha curcas] |
| 6 |
Hb_002675_030 |
0.0720657052 |
- |
- |
coatomer beta subunit, putative [Ricinus communis] |
| 7 |
Hb_001329_250 |
0.0774622923 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105642692 isoform X1 [Jatropha curcas] |
| 8 |
Hb_004102_170 |
0.0785711437 |
- |
- |
PREDICTED: lys-63-specific deubiquitinase BRCC36-like [Jatropha curcas] |
| 9 |
Hb_009569_040 |
0.0796316983 |
- |
- |
PREDICTED: uncharacterized protein LOC105635573 [Jatropha curcas] |
| 10 |
Hb_000049_020 |
0.0811154427 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 11 |
Hb_003498_080 |
0.0836834278 |
- |
- |
PREDICTED: decapping 5-like protein isoform X2 [Jatropha curcas] |
| 12 |
Hb_012055_040 |
0.0839427666 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 13 |
Hb_001821_150 |
0.0839803728 |
- |
- |
PREDICTED: potassium transporter 7-like [Jatropha curcas] |
| 14 |
Hb_003417_010 |
0.0841215921 |
- |
- |
PREDICTED: protein TPLATE [Jatropha curcas] |
| 15 |
Hb_000046_500 |
0.0860621059 |
- |
- |
PREDICTED: probable serine/threonine-protein phosphatase 2A regulatory subunit B'' subunit TON2 [Jatropha curcas] |
| 16 |
Hb_004679_030 |
0.0861073612 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: F-box protein SKIP23-like [Jatropha curcas] |
| 17 |
Hb_000212_420 |
0.0861613166 |
- |
- |
PREDICTED: probable protein phosphatase 2C 33 [Jatropha curcas] |
| 18 |
Hb_000773_040 |
0.0864095374 |
- |
- |
PREDICTED: uncharacterized protein LOC105641863 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000960_040 |
0.0864259978 |
- |
- |
PREDICTED: ERAD-associated E3 ubiquitin-protein ligase HRD1B-like [Vitis vinifera] |
| 20 |
Hb_159809_030 |
0.0865961823 |
- |
- |
PREDICTED: UDP-D-xylose:L-fucose alpha-1,3-D-xylosyltransferase 2-like isoform X2 [Jatropha curcas] |