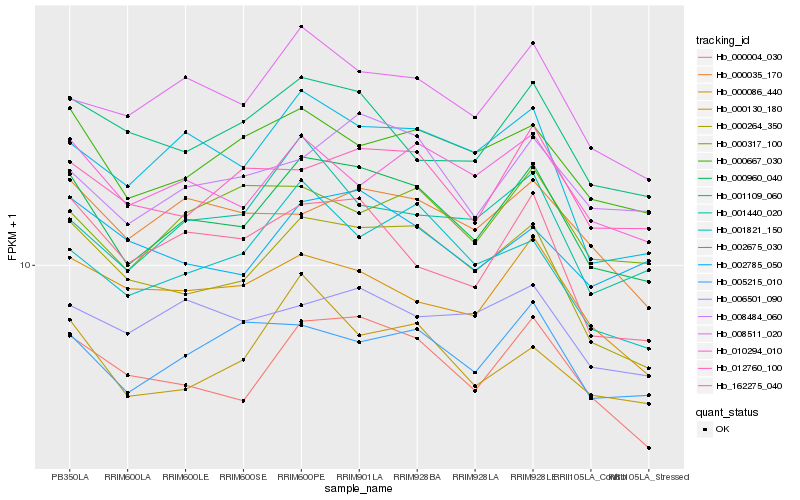
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000960_040 |
0.0 |
- |
- |
PREDICTED: ERAD-associated E3 ubiquitin-protein ligase HRD1B-like [Vitis vinifera] |
| 2 |
Hb_000004_030 |
0.065301764 |
- |
- |
PREDICTED: Niemann-Pick C1 protein [Jatropha curcas] |
| 3 |
Hb_000264_350 |
0.0664342325 |
- |
- |
PREDICTED: uncharacterized protein LOC105649541 [Jatropha curcas] |
| 4 |
Hb_008511_020 |
0.0725949348 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 5 |
Hb_000667_030 |
0.0742433137 |
- |
- |
PREDICTED: protein transport protein SEC23-like [Jatropha curcas] |
| 6 |
Hb_002675_030 |
0.0776194505 |
- |
- |
coatomer beta subunit, putative [Ricinus communis] |
| 7 |
Hb_010294_010 |
0.0785779478 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3B [Jatropha curcas] |
| 8 |
Hb_005215_010 |
0.0802021948 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_008484_060 |
0.0806638718 |
- |
- |
PREDICTED: allantoate deiminase isoform X3 [Populus euphratica] |
| 10 |
Hb_012760_100 |
0.0825731158 |
- |
- |
Pre-rRNA-processing protein ESF1, putative [Ricinus communis] |
| 11 |
Hb_001109_060 |
0.0835835883 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 12 |
Hb_162275_040 |
0.0837776977 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000130_180 |
0.0851640969 |
- |
- |
PREDICTED: GPI inositol-deacylase-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_000086_440 |
0.0854284851 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |
| 15 |
Hb_001440_020 |
0.0864259978 |
- |
- |
exocyst componenet sec8, putative [Ricinus communis] |
| 16 |
Hb_001821_150 |
0.0885983371 |
- |
- |
PREDICTED: potassium transporter 7-like [Jatropha curcas] |
| 17 |
Hb_000035_170 |
0.0889487807 |
- |
- |
PREDICTED: DNA-directed RNA polymerases IV and V subunit 2-like [Jatropha curcas] |
| 18 |
Hb_006501_090 |
0.0892176483 |
- |
- |
PREDICTED: golgin candidate 1 [Jatropha curcas] |
| 19 |
Hb_002785_050 |
0.0894972254 |
- |
- |
PREDICTED: adenylosuccinate lyase-like [Jatropha curcas] |
| 20 |
Hb_000317_100 |
0.0903542701 |
- |
- |
PREDICTED: uncharacterized protein LOC105646995 [Jatropha curcas] |