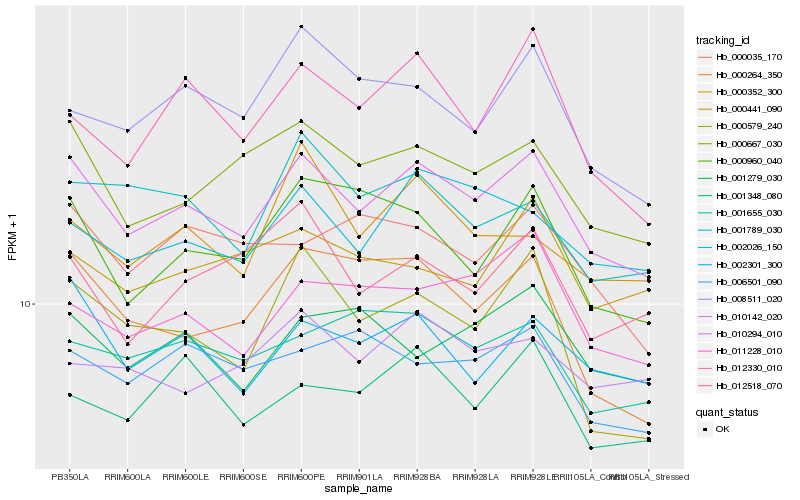
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010294_010 |
0.0 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit STT3B [Jatropha curcas] |
| 2 |
Hb_000667_030 |
0.0634225612 |
- |
- |
PREDICTED: protein transport protein SEC23-like [Jatropha curcas] |
| 3 |
Hb_002301_300 |
0.0704991026 |
- |
- |
PREDICTED: dol-P-Glc:Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase isoform X1 [Jatropha curcas] |
| 4 |
Hb_002026_150 |
0.0766750035 |
transcription factor |
TF Family: SNF2 |
PREDICTED: DNA repair helicase XPB1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_011228_010 |
0.0774750777 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 54, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_000960_040 |
0.0785779478 |
- |
- |
PREDICTED: ERAD-associated E3 ubiquitin-protein ligase HRD1B-like [Vitis vinifera] |
| 7 |
Hb_012330_010 |
0.0794254055 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 12-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_001348_080 |
0.0823794332 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 9 |
Hb_001655_030 |
0.08274595 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 3 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000264_350 |
0.0827959279 |
- |
- |
PREDICTED: uncharacterized protein LOC105649541 [Jatropha curcas] |
| 11 |
Hb_008511_020 |
0.0829681404 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 12 |
Hb_000035_170 |
0.0840271373 |
- |
- |
PREDICTED: DNA-directed RNA polymerases IV and V subunit 2-like [Jatropha curcas] |
| 13 |
Hb_012518_070 |
0.0845291684 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 9 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001789_030 |
0.0850934811 |
- |
- |
PREDICTED: signal peptide peptidase-like 1 [Jatropha curcas] |
| 15 |
Hb_001279_030 |
0.0852188987 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' theta isoform-like [Jatropha curcas] |
| 16 |
Hb_006501_090 |
0.0853940596 |
- |
- |
PREDICTED: golgin candidate 1 [Jatropha curcas] |
| 17 |
Hb_000579_240 |
0.0855285621 |
- |
- |
phosphatidylinositol transporter, putative [Ricinus communis] |
| 18 |
Hb_010142_020 |
0.0860845152 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000441_090 |
0.0863757706 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory subunit 2 [Jatropha curcas] |
| 20 |
Hb_000352_300 |
0.0864724121 |
- |
- |
PREDICTED: eukaryotic peptide chain release factor subunit 1-3 [Jatropha curcas] |