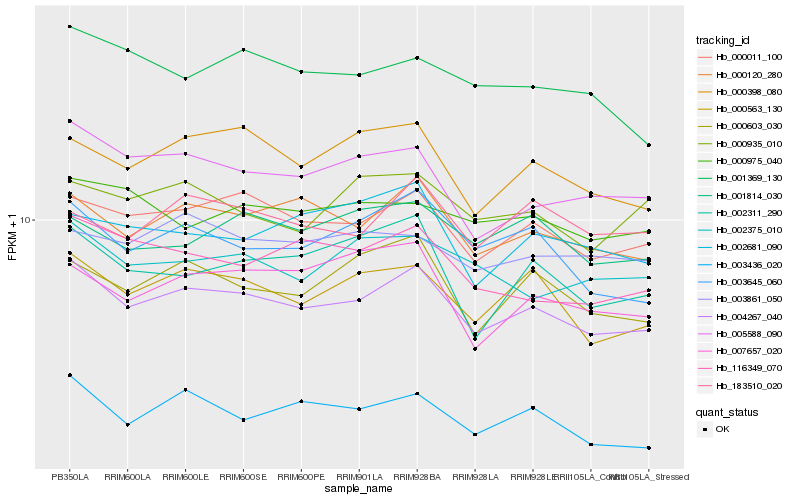
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004267_040 |
0.0 |
- |
- |
PREDICTED: transcriptional adapter ADA2a [Jatropha curcas] |
| 2 |
Hb_000011_100 |
0.0446715111 |
- |
- |
PREDICTED: uncharacterized protein C630.12 [Jatropha curcas] |
| 3 |
Hb_000120_280 |
0.0486933041 |
- |
- |
PREDICTED: protein transport protein SEC23 [Jatropha curcas] |
| 4 |
Hb_000563_130 |
0.0518571062 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 5 |
Hb_005588_090 |
0.0571932077 |
- |
- |
hypothetical protein CICLE_v10003480mg [Citrus clementina] |
| 6 |
Hb_000398_080 |
0.0586951249 |
- |
- |
tip120, putative [Ricinus communis] |
| 7 |
Hb_000603_030 |
0.0613262895 |
- |
- |
Ribonuclease III, putative [Ricinus communis] |
| 8 |
Hb_003645_060 |
0.0613298974 |
- |
- |
PREDICTED: monodehydroascorbate reductase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_116349_070 |
0.0657984622 |
- |
- |
hypothetical protein JCGZ_21753 [Jatropha curcas] |
| 10 |
Hb_002311_290 |
0.0683546779 |
- |
- |
PREDICTED: pumilio homolog 24 [Jatropha curcas] |
| 11 |
Hb_002375_010 |
0.0689827745 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X1 [Jatropha curcas] |
| 12 |
Hb_003436_020 |
0.0691265048 |
transcription factor |
TF Family: Orphans |
PREDICTED: uncharacterized protein LOC105636276 isoform X2 [Jatropha curcas] |
| 13 |
Hb_007657_020 |
0.0704099694 |
- |
- |
PREDICTED: PRA1 family protein H isoform X1 [Jatropha curcas] |
| 14 |
Hb_183510_020 |
0.0708316836 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 15 |
Hb_001369_130 |
0.0712097219 |
- |
- |
PREDICTED: autophagy-related protein 3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_003861_050 |
0.0718185373 |
- |
- |
PREDICTED: uncharacterized protein LOC105650779 [Jatropha curcas] |
| 17 |
Hb_001814_030 |
0.0728884574 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |
| 18 |
Hb_002681_090 |
0.0730117845 |
- |
- |
PREDICTED: golgin candidate 6 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000935_010 |
0.0732733148 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 16 [Jatropha curcas] |
| 20 |
Hb_000975_040 |
0.0740286263 |
- |
- |
hypothetical protein CICLE_v10028249mg [Citrus clementina] |