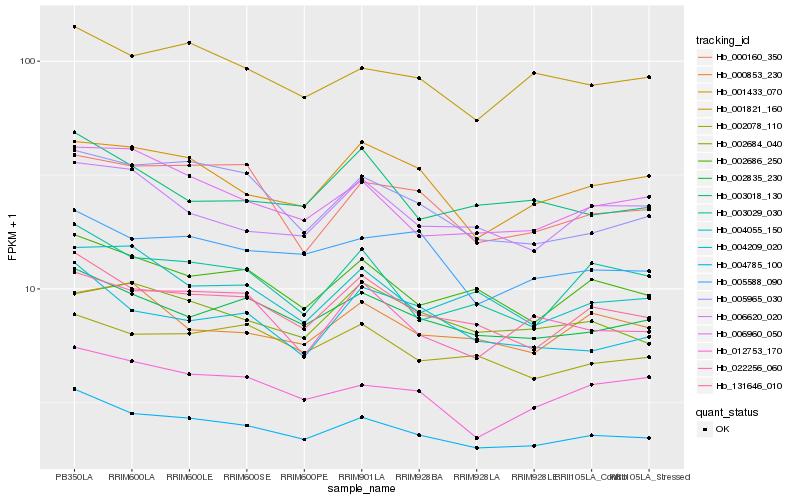
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004785_100 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_131646_010 |
0.0386568894 |
- |
- |
protein arginine n-methyltransferase, putative [Ricinus communis] |
| 3 |
Hb_002835_230 |
0.0431762993 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_002686_250 |
0.0520389229 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 65 kDa protein isoform X1 [Jatropha curcas] |
| 5 |
Hb_001821_160 |
0.0556876751 |
- |
- |
serine/arginine rich splicing factor, putative [Ricinus communis] |
| 6 |
Hb_006960_050 |
0.0590131738 |
- |
- |
PREDICTED: PRKR-interacting protein 1 [Jatropha curcas] |
| 7 |
Hb_002078_110 |
0.0606694894 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 8 |
Hb_005588_090 |
0.0617272546 |
- |
- |
hypothetical protein CICLE_v10003480mg [Citrus clementina] |
| 9 |
Hb_005965_030 |
0.0621118909 |
- |
- |
hypothetical protein JCGZ_16415 [Jatropha curcas] |
| 10 |
Hb_012753_170 |
0.0664273418 |
- |
- |
PREDICTED: phosphatidylinositol glycan anchor biosynthesis class U protein-like isoform X2 [Jatropha curcas] |
| 11 |
Hb_004055_150 |
0.0675210253 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004209_020 |
0.0679497671 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 51-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_022256_060 |
0.0687116407 |
- |
- |
PREDICTED: uncharacterized protein LOC105628137 [Jatropha curcas] |
| 14 |
Hb_003018_130 |
0.0703733813 |
- |
- |
PREDICTED: uncharacterized protein LOC105628777 [Jatropha curcas] |
| 15 |
Hb_006620_020 |
0.0706533012 |
- |
- |
PREDICTED: uncharacterized protein LOC105638878 [Jatropha curcas] |
| 16 |
Hb_003029_030 |
0.0709897826 |
- |
- |
PREDICTED: uncharacterized protein LOC105642632 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001433_070 |
0.0712681797 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1 [Jatropha curcas] |
| 18 |
Hb_000160_350 |
0.0737656747 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 4 isoform X3 [Vitis vinifera] |
| 19 |
Hb_000853_230 |
0.0760982987 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 20 |
Hb_002684_040 |
0.0761427101 |
- |
- |
PREDICTED: uncharacterized protein LOC105634392 isoform X1 [Jatropha curcas] |