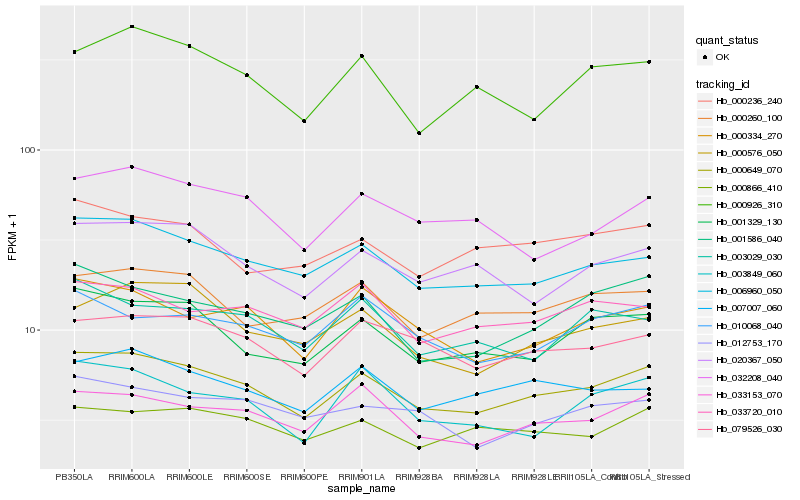
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000649_070 |
0.0 |
- |
- |
PREDICTED: probable UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase 2 [Jatropha curcas] |
| 2 |
Hb_006960_050 |
0.0553825187 |
- |
- |
PREDICTED: PRKR-interacting protein 1 [Jatropha curcas] |
| 3 |
Hb_001329_130 |
0.0638336981 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_033153_070 |
0.0663383879 |
- |
- |
PREDICTED: uncharacterized protein LOC105649606 isoform X1 [Jatropha curcas] |
| 5 |
Hb_079526_030 |
0.0686181332 |
- |
- |
PREDICTED: uncharacterized protein LOC105638120 [Jatropha curcas] |
| 6 |
Hb_000334_270 |
0.072127835 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_020367_050 |
0.0722146119 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranylgeranyl pyrophosphate synthase, putative [Ricinus communis] |
| 8 |
Hb_007007_060 |
0.0737526562 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_000260_100 |
0.074881574 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000866_410 |
0.0760433364 |
- |
- |
hypothetical protein JCGZ_14422 [Jatropha curcas] |
| 11 |
Hb_001586_040 |
0.0764241423 |
- |
- |
hypothetical protein JCGZ_17187 [Jatropha curcas] |
| 12 |
Hb_000576_050 |
0.0772659607 |
- |
- |
Kinase superfamily protein [Theobroma cacao] |
| 13 |
Hb_012753_170 |
0.0777780166 |
- |
- |
PREDICTED: phosphatidylinositol glycan anchor biosynthesis class U protein-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_000926_310 |
0.0781760953 |
- |
- |
uncharacterized protein LOC100500241 [Glycine max] |
| 15 |
Hb_032208_040 |
0.0800949256 |
- |
- |
PREDICTED: serine/arginine-rich SC35-like splicing factor SCL33 [Populus euphratica] |
| 16 |
Hb_000236_240 |
0.0806544624 |
- |
- |
Pre-mRNA-splicing factor cwc15, putative [Ricinus communis] |
| 17 |
Hb_010068_040 |
0.0808151126 |
- |
- |
PREDICTED: uncharacterized protein LOC105632964 [Jatropha curcas] |
| 18 |
Hb_003029_030 |
0.080961159 |
- |
- |
PREDICTED: uncharacterized protein LOC105642632 isoform X2 [Jatropha curcas] |
| 19 |
Hb_033720_010 |
0.0814550171 |
- |
- |
PREDICTED: uncharacterized protein LOC105648843 [Jatropha curcas] |
| 20 |
Hb_003849_060 |
0.0828527868 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g13040, mitochondrial [Jatropha curcas] |