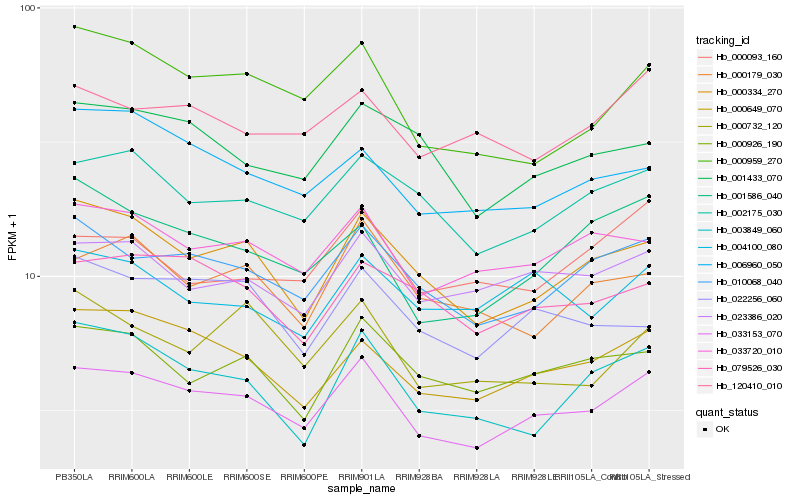
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033153_070 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649606 isoform X1 [Jatropha curcas] |
| 2 |
Hb_010068_040 |
0.06004447 |
- |
- |
PREDICTED: uncharacterized protein LOC105632964 [Jatropha curcas] |
| 3 |
Hb_000334_270 |
0.064889978 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000649_070 |
0.0663383879 |
- |
- |
PREDICTED: probable UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase 2 [Jatropha curcas] |
| 5 |
Hb_033720_010 |
0.0710054737 |
- |
- |
PREDICTED: uncharacterized protein LOC105648843 [Jatropha curcas] |
| 6 |
Hb_002175_030 |
0.0719334581 |
- |
- |
PREDICTED: snRNA-activating protein complex subunit isoform X2 [Jatropha curcas] |
| 7 |
Hb_000959_270 |
0.0725387707 |
- |
- |
PREDICTED: uncharacterized protein LOC105641005 [Jatropha curcas] |
| 8 |
Hb_023386_020 |
0.0739368509 |
- |
- |
PREDICTED: chaperone protein dnaJ 13 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000926_190 |
0.0780768248 |
- |
- |
PREDICTED: 1,4-alpha-glucan-branching enzyme 3, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 10 |
Hb_001586_040 |
0.0813470217 |
- |
- |
hypothetical protein JCGZ_17187 [Jatropha curcas] |
| 11 |
Hb_079526_030 |
0.0818980667 |
- |
- |
PREDICTED: uncharacterized protein LOC105638120 [Jatropha curcas] |
| 12 |
Hb_006960_050 |
0.0830965133 |
- |
- |
PREDICTED: PRKR-interacting protein 1 [Jatropha curcas] |
| 13 |
Hb_000179_030 |
0.0833441661 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001433_070 |
0.0848246541 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1 [Jatropha curcas] |
| 15 |
Hb_120410_010 |
0.0853817769 |
- |
- |
PREDICTED: vesicle-associated membrane protein 727-like [Solanum tuberosum] |
| 16 |
Hb_003849_060 |
0.0856936891 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g13040, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000732_120 |
0.0869437039 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 18 |
Hb_022256_060 |
0.087405881 |
- |
- |
PREDICTED: uncharacterized protein LOC105628137 [Jatropha curcas] |
| 19 |
Hb_004100_080 |
0.0875918 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000093_160 |
0.0881704012 |
- |
- |
PREDICTED: nuclear transcription factor Y subunit B-1-like isoform X1 [Jatropha curcas] |