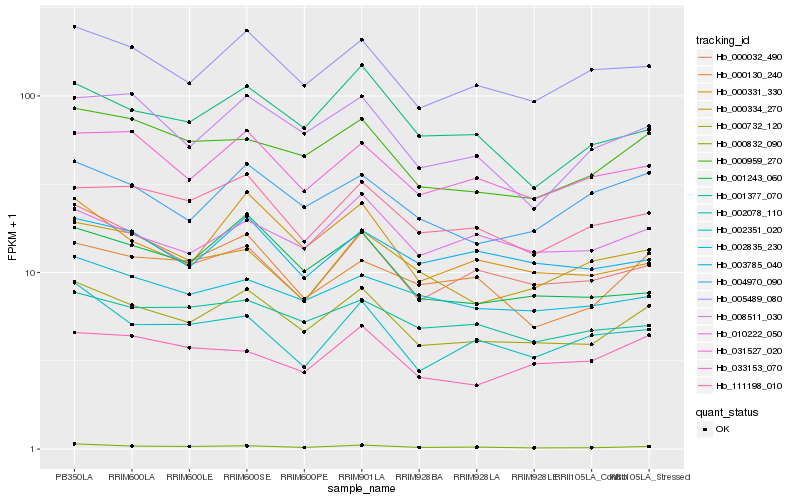
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000732_120 |
0.0 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 2 |
Hb_005489_080 |
0.059136436 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000959_270 |
0.0681608085 |
- |
- |
PREDICTED: uncharacterized protein LOC105641005 [Jatropha curcas] |
| 4 |
Hb_000832_090 |
0.0698799808 |
transcription factor |
TF Family: mTERF |
hypothetical protein VITISV_028994 [Vitis vinifera] |
| 5 |
Hb_031527_020 |
0.0699597579 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B isoform X1 [Jatropha curcas] |
| 6 |
Hb_111198_010 |
0.0735903905 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 37-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_004970_090 |
0.0779081335 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 2 [Jatropha curcas] |
| 8 |
Hb_002835_230 |
0.0815901567 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_008511_030 |
0.0821065101 |
- |
- |
guanine nucleotide-binding protein beta, putative [Ricinus communis] |
| 10 |
Hb_010222_050 |
0.0828761607 |
- |
- |
PREDICTED: WD repeat-containing protein 48 isoform X2 [Jatropha curcas] |
| 11 |
Hb_001377_070 |
0.0837747279 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003785_040 |
0.0841847432 |
- |
- |
PREDICTED: R3H domain-containing protein 2-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000032_490 |
0.0850984493 |
- |
- |
PREDICTED: uncharacterized protein At4g10930-like [Jatropha curcas] |
| 14 |
Hb_000331_330 |
0.0854393648 |
- |
- |
PREDICTED: transcription factor GTE10 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002078_110 |
0.0861708878 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 16 |
Hb_033153_070 |
0.0869437039 |
- |
- |
PREDICTED: uncharacterized protein LOC105649606 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000130_240 |
0.0871696369 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 18 |
Hb_000334_270 |
0.0882954286 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_001243_060 |
0.0886513812 |
- |
- |
PREDICTED: probable magnesium transporter NIPA6 [Jatropha curcas] |
| 20 |
Hb_002351_020 |
0.0888875665 |
- |
- |
PREDICTED: rRNA methyltransferase 3B, mitochondrial [Jatropha curcas] |