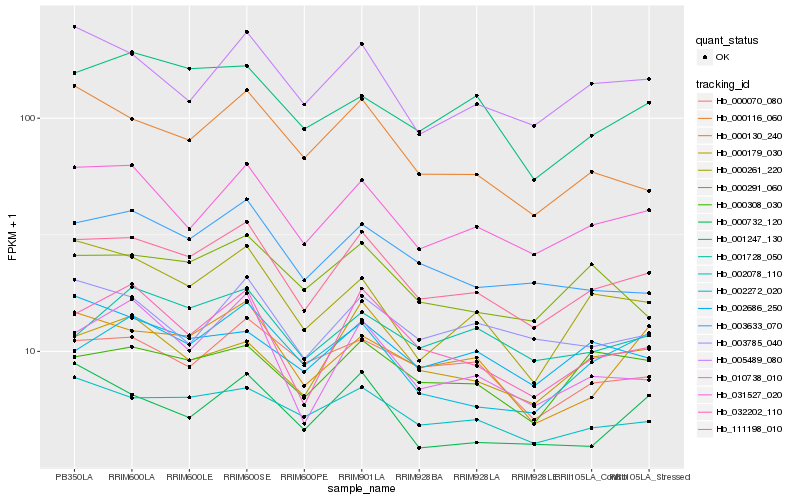
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_111198_010 |
0.0 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 37-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_031527_020 |
0.0544318265 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B isoform X1 [Jatropha curcas] |
| 3 |
Hb_032202_110 |
0.0599215975 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 48-like [Jatropha curcas] |
| 4 |
Hb_003633_070 |
0.0675755001 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |
| 5 |
Hb_002078_110 |
0.0681646361 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 6 |
Hb_005489_080 |
0.0684600651 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_010738_010 |
0.0697076318 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 8 |
Hb_000130_240 |
0.0722425285 |
- |
- |
hypothetical protein RCOM_1520720 [Ricinus communis] |
| 9 |
Hb_000070_080 |
0.0722868446 |
- |
- |
PREDICTED: uncharacterized protein LOC105645742 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000308_030 |
0.0732258974 |
- |
- |
PREDICTED: uncharacterized protein LOC105639686 [Jatropha curcas] |
| 11 |
Hb_000179_030 |
0.0735025049 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000732_120 |
0.0735903905 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000291_060 |
0.0750055148 |
transcription factor |
TF Family: C2H2 |
PREDICTED: MYST-like histone acetyltransferase 2 [Jatropha curcas] |
| 14 |
Hb_000116_060 |
0.0754294324 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 15 |
Hb_000261_220 |
0.0763721579 |
- |
- |
PREDICTED: TBC1 domain family member 13 [Jatropha curcas] |
| 16 |
Hb_002272_020 |
0.0764304988 |
transcription factor |
TF Family: NF-YC |
PREDICTED: nuclear transcription factor Y subunit C-3 [Jatropha curcas] |
| 17 |
Hb_003785_040 |
0.0764609736 |
- |
- |
PREDICTED: R3H domain-containing protein 2-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_002686_250 |
0.0770246206 |
- |
- |
PREDICTED: U11/U12 small nuclear ribonucleoprotein 65 kDa protein isoform X1 [Jatropha curcas] |
| 19 |
Hb_001247_130 |
0.0771634843 |
- |
- |
PREDICTED: shaggy-related protein kinase alpha [Jatropha curcas] |
| 20 |
Hb_001728_050 |
0.077440286 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RBBP6-like [Jatropha curcas] |