| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
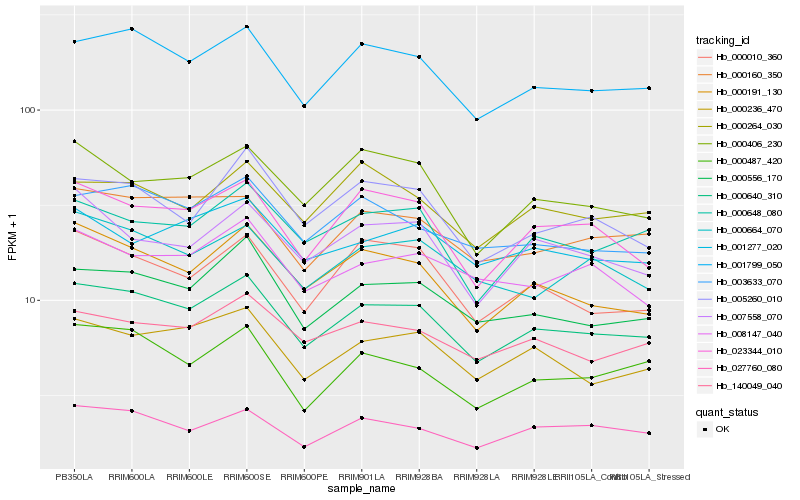
Hb_000640_310 |
0.0 |
- |
- |
hypothetical protein JCGZ_14055 [Jatropha curcas] |
| 2 |
Hb_001799_050 |
0.0369412487 |
- |
- |
PREDICTED: plasminogen activator inhibitor 1 RNA-binding protein [Vitis vinifera] |
| 3 |
Hb_000556_170 |
0.0525301074 |
- |
- |
PREDICTED: uncharacterized protein LOC105643102 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000236_470 |
0.0575500151 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g55840 [Jatropha curcas] |
| 5 |
Hb_000191_130 |
0.0576859713 |
- |
- |
PREDICTED: RNA polymerase-associated protein CTR9 homolog [Jatropha curcas] |
| 6 |
Hb_000160_350 |
0.060451158 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 4 isoform X3 [Vitis vinifera] |
| 7 |
Hb_023344_010 |
0.0608700296 |
- |
- |
PREDICTED: pre-mRNA-processing factor 39 isoform X2 [Jatropha curcas] |
| 8 |
Hb_003633_070 |
0.061689125 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |
| 9 |
Hb_140049_040 |
0.0622557526 |
- |
- |
PREDICTED: uncharacterized protein LOC105632012 [Jatropha curcas] |
| 10 |
Hb_027760_080 |
0.0626887643 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 11 |
Hb_000648_080 |
0.0649109056 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 1 isoform X2 [Populus euphratica] |
| 12 |
Hb_000010_360 |
0.0677340783 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY [Jatropha curcas] |
| 13 |
Hb_005260_010 |
0.0683627061 |
- |
- |
PREDICTED: golgin candidate 6 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000406_230 |
0.0686687007 |
- |
- |
PREDICTED: probable nucleolar protein 5-1 [Jatropha curcas] |
| 15 |
Hb_000664_070 |
0.0689667017 |
- |
- |
PREDICTED: protein cereblon [Jatropha curcas] |
| 16 |
Hb_000487_420 |
0.0692861144 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At1g04390 [Jatropha curcas] |
| 17 |
Hb_007558_070 |
0.0725469547 |
- |
- |
PREDICTED: pre-mRNA-processing factor 39 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000264_030 |
0.072551454 |
- |
- |
PREDICTED: splicing factor 3B subunit 2 [Jatropha curcas] |
| 19 |
Hb_001277_020 |
0.0727531094 |
- |
- |
PREDICTED: armadillo repeat-containing protein LFR [Jatropha curcas] |
| 20 |
Hb_008147_040 |
0.0741323788 |
- |
- |
PREDICTED: protein FLX-like 2 [Jatropha curcas] |