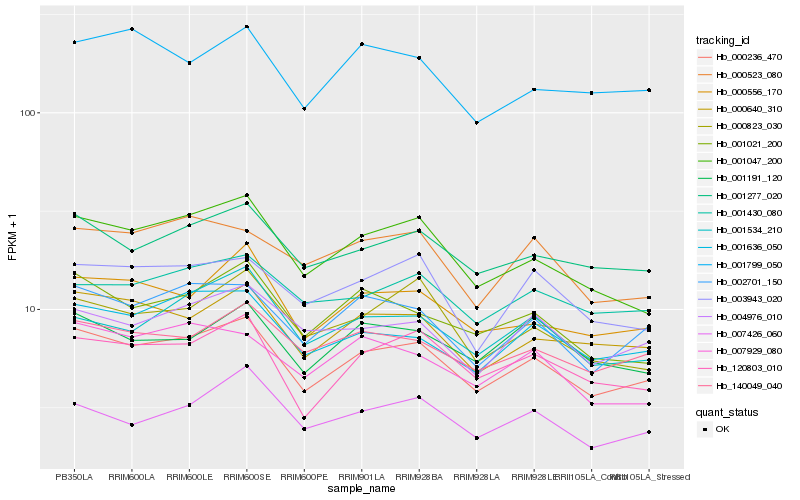
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000236_470 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g55840 [Jatropha curcas] |
| 2 |
Hb_001277_020 |
0.056736097 |
- |
- |
PREDICTED: armadillo repeat-containing protein LFR [Jatropha curcas] |
| 3 |
Hb_000640_310 |
0.0575500151 |
- |
- |
hypothetical protein JCGZ_14055 [Jatropha curcas] |
| 4 |
Hb_002701_150 |
0.0576448919 |
- |
- |
PREDICTED: probable tRNA (guanine(26)-N(2))-dimethyltransferase 2 isoform X2 [Jatropha curcas] |
| 5 |
Hb_001047_200 |
0.0604542765 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 18 [Eucalyptus grandis] |
| 6 |
Hb_120803_010 |
0.0617636742 |
- |
- |
PREDICTED: protein kinase and PP2C-like domain-containing protein [Malus domestica] |
| 7 |
Hb_000556_170 |
0.0622554948 |
- |
- |
PREDICTED: uncharacterized protein LOC105643102 isoform X2 [Jatropha curcas] |
| 8 |
Hb_001636_050 |
0.0623765514 |
transcription factor |
TF Family: NF-X1 |
nuclear transcription factor, X-box binding, putative [Ricinus communis] |
| 9 |
Hb_140049_040 |
0.0652098517 |
- |
- |
PREDICTED: uncharacterized protein LOC105632012 [Jatropha curcas] |
| 10 |
Hb_001021_200 |
0.0685976953 |
desease resistance |
Gene Name: AAA |
PREDICTED: peroxisome biogenesis protein 6 [Jatropha curcas] |
| 11 |
Hb_004976_010 |
0.0704961263 |
- |
- |
hypothetical protein RCOM_1429110 [Ricinus communis] |
| 12 |
Hb_001430_080 |
0.0723728619 |
- |
- |
PREDICTED: putative mediator of RNA polymerase II transcription subunit 26 [Jatropha curcas] |
| 13 |
Hb_003943_020 |
0.0737387835 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase PASTICCINO1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_007426_060 |
0.0747049259 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Coffea canephora] |
| 15 |
Hb_001534_210 |
0.0751787418 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105642316 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000823_030 |
0.0758483205 |
- |
- |
PREDICTED: DNA-directed RNA polymerase III subunit RPC5 [Jatropha curcas] |
| 17 |
Hb_001191_120 |
0.0759194887 |
- |
- |
PREDICTED: cleavage stimulating factor 64 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000523_080 |
0.0761137544 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: beta-catenin-like protein 1 [Jatropha curcas] |
| 19 |
Hb_001799_050 |
0.0773368636 |
- |
- |
PREDICTED: plasminogen activator inhibitor 1 RNA-binding protein [Vitis vinifera] |
| 20 |
Hb_007929_080 |
0.0777497546 |
- |
- |
PREDICTED: transforming growth factor-beta receptor-associated protein 1 [Jatropha curcas] |