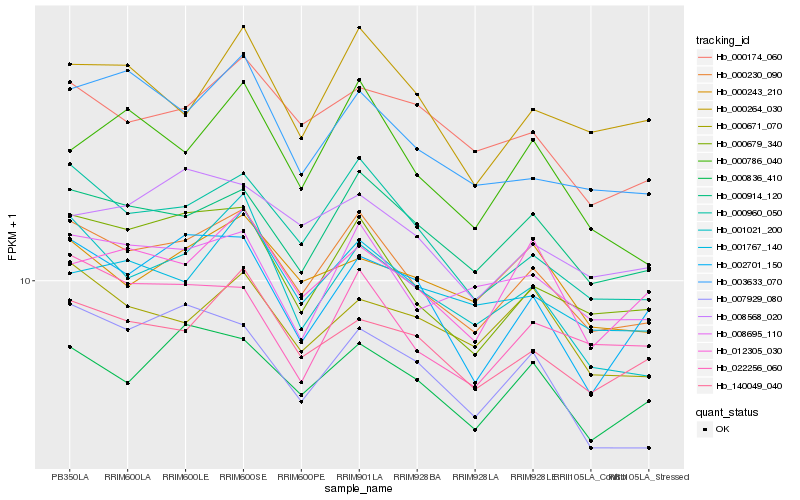
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000230_090 |
0.0 |
- |
- |
PREDICTED: WD repeat-containing protein 48 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000960_050 |
0.0410232246 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |
| 3 |
Hb_000914_120 |
0.0463163406 |
- |
- |
PREDICTED: outer envelope protein 80, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000679_340 |
0.0497466638 |
- |
- |
PREDICTED: uncharacterized protein LOC105650070 isoform X1 [Jatropha curcas] |
| 5 |
Hb_140049_040 |
0.0588479821 |
- |
- |
PREDICTED: uncharacterized protein LOC105632012 [Jatropha curcas] |
| 6 |
Hb_000836_410 |
0.0590711742 |
- |
- |
sec10, putative [Ricinus communis] |
| 7 |
Hb_008695_110 |
0.061378966 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105638874 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000174_060 |
0.0618370275 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 9 |
Hb_001021_200 |
0.0637460722 |
desease resistance |
Gene Name: AAA |
PREDICTED: peroxisome biogenesis protein 6 [Jatropha curcas] |
| 10 |
Hb_022256_060 |
0.0646111637 |
- |
- |
PREDICTED: uncharacterized protein LOC105628137 [Jatropha curcas] |
| 11 |
Hb_001767_140 |
0.0655310549 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |
| 12 |
Hb_003633_070 |
0.0656775128 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |
| 13 |
Hb_000264_030 |
0.0667013721 |
- |
- |
PREDICTED: splicing factor 3B subunit 2 [Jatropha curcas] |
| 14 |
Hb_008568_020 |
0.0673188015 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |
| 15 |
Hb_000671_070 |
0.069393117 |
- |
- |
PREDICTED: uncharacterized protein LOC105630621 [Jatropha curcas] |
| 16 |
Hb_000243_210 |
0.0697953722 |
- |
- |
protein with unknown function [Ricinus communis] |
| 17 |
Hb_012305_030 |
0.0700241566 |
- |
- |
hypothetical protein JCGZ_18979 [Jatropha curcas] |
| 18 |
Hb_007929_080 |
0.070509246 |
- |
- |
PREDICTED: transforming growth factor-beta receptor-associated protein 1 [Jatropha curcas] |
| 19 |
Hb_002701_150 |
0.0706363239 |
- |
- |
PREDICTED: probable tRNA (guanine(26)-N(2))-dimethyltransferase 2 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000786_040 |
0.0710117362 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 17 [Jatropha curcas] |