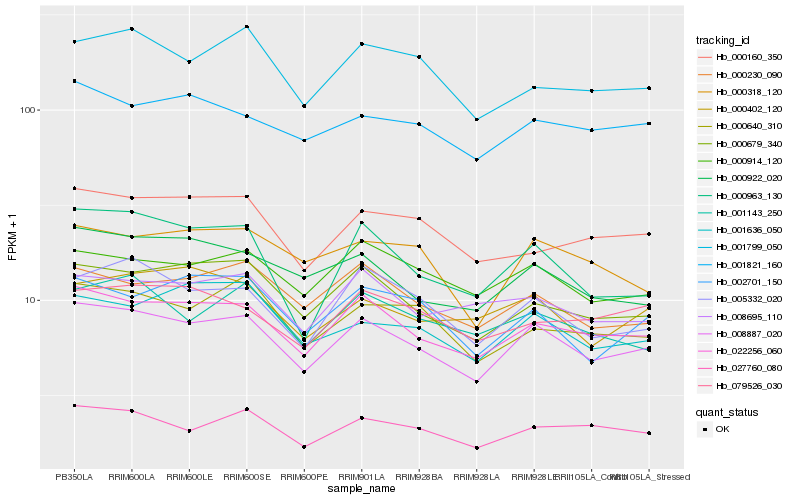
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008887_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636191 [Jatropha curcas] |
| 2 |
Hb_022256_060 |
0.0443181312 |
- |
- |
PREDICTED: uncharacterized protein LOC105628137 [Jatropha curcas] |
| 3 |
Hb_000679_340 |
0.0653110346 |
- |
- |
PREDICTED: uncharacterized protein LOC105650070 isoform X1 [Jatropha curcas] |
| 4 |
Hb_027760_080 |
0.0680189792 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 5 |
Hb_005332_020 |
0.0707760974 |
transcription factor |
TF Family: SNF2 |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 6 |
Hb_000914_120 |
0.0717857768 |
- |
- |
PREDICTED: outer envelope protein 80, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000230_090 |
0.0734508131 |
- |
- |
PREDICTED: WD repeat-containing protein 48 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000922_020 |
0.0738858907 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 9 |
Hb_001821_160 |
0.0764559904 |
- |
- |
serine/arginine rich splicing factor, putative [Ricinus communis] |
| 10 |
Hb_000640_310 |
0.077550766 |
- |
- |
hypothetical protein JCGZ_14055 [Jatropha curcas] |
| 11 |
Hb_008695_110 |
0.0796306922 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105638874 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001799_050 |
0.0798540099 |
- |
- |
PREDICTED: plasminogen activator inhibitor 1 RNA-binding protein [Vitis vinifera] |
| 13 |
Hb_001143_250 |
0.0800344937 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3C isoform X1 [Jatropha curcas] |
| 14 |
Hb_002701_150 |
0.0803064662 |
- |
- |
PREDICTED: probable tRNA (guanine(26)-N(2))-dimethyltransferase 2 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001636_050 |
0.081264941 |
transcription factor |
TF Family: NF-X1 |
nuclear transcription factor, X-box binding, putative [Ricinus communis] |
| 16 |
Hb_000160_350 |
0.0819261208 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 4 isoform X3 [Vitis vinifera] |
| 17 |
Hb_000402_120 |
0.0821430439 |
- |
- |
PREDICTED: uncharacterized protein LOC105634656 [Jatropha curcas] |
| 18 |
Hb_000963_130 |
0.0825293162 |
- |
- |
RNA m5u methyltransferase, putative [Ricinus communis] |
| 19 |
Hb_000318_120 |
0.0835871683 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: DNA ligase 1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_079526_030 |
0.083849776 |
- |
- |
PREDICTED: uncharacterized protein LOC105638120 [Jatropha curcas] |