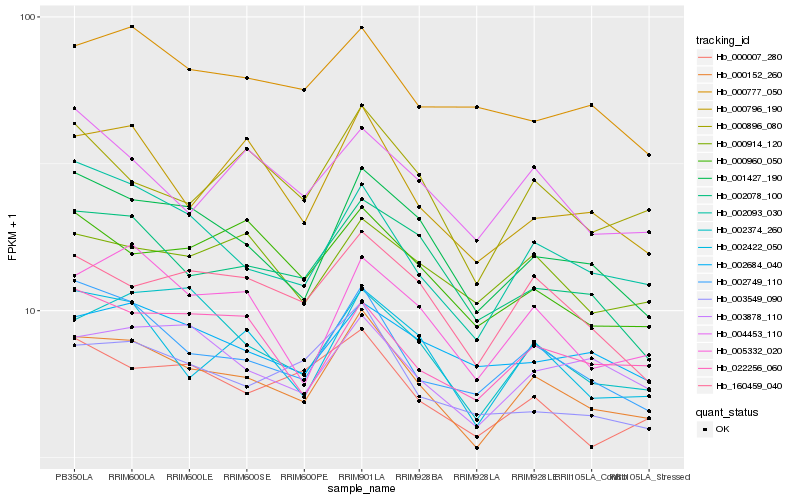
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000152_260 |
0.0 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 1 [Jatropha curcas] |
| 2 |
Hb_002749_110 |
0.0609602407 |
- |
- |
PREDICTED: uncharacterized protein LOC105636001 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001427_190 |
0.065158798 |
- |
- |
PREDICTED: histone acetyltransferase type B catalytic subunit [Jatropha curcas] |
| 4 |
Hb_002422_050 |
0.0667472372 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 5 isoform X1 [Jatropha curcas] |
| 5 |
Hb_160459_040 |
0.0694906861 |
- |
- |
hypothetical protein JCGZ_01511 [Jatropha curcas] |
| 6 |
Hb_002374_260 |
0.0700835782 |
- |
- |
PREDICTED: probable methyltransferase-like protein 15 [Jatropha curcas] |
| 7 |
Hb_003878_110 |
0.07098717 |
- |
- |
PREDICTED: uncharacterized protein LOC105637024 [Jatropha curcas] |
| 8 |
Hb_002093_030 |
0.071608783 |
- |
- |
PREDICTED: uncharacterized protein LOC105648523 [Jatropha curcas] |
| 9 |
Hb_005332_020 |
0.0717388853 |
transcription factor |
TF Family: SNF2 |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 10 |
Hb_000007_280 |
0.0726029772 |
- |
- |
hypothetical protein VITISV_000066 [Vitis vinifera] |
| 11 |
Hb_002078_100 |
0.0734347801 |
- |
- |
PREDICTED: mannosyl-oligosaccharide glucosidase GCS1-like [Jatropha curcas] |
| 12 |
Hb_022256_060 |
0.0743802009 |
- |
- |
PREDICTED: uncharacterized protein LOC105628137 [Jatropha curcas] |
| 13 |
Hb_000914_120 |
0.0745821499 |
- |
- |
PREDICTED: outer envelope protein 80, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000896_080 |
0.0753638918 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP95 isoform X1 [Vitis vinifera] |
| 15 |
Hb_003549_090 |
0.0769044988 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 6 [Jatropha curcas] |
| 16 |
Hb_002684_040 |
0.0779295541 |
- |
- |
PREDICTED: uncharacterized protein LOC105634392 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000796_190 |
0.0779464453 |
- |
- |
hypothetical protein F383_31148 [Gossypium arboreum] |
| 18 |
Hb_000960_050 |
0.0798148206 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |
| 19 |
Hb_004453_110 |
0.0809402274 |
- |
- |
Peptidyl-prolyl cis-trans isomerase CYP19-2 isoform 1 [Theobroma cacao] |
| 20 |
Hb_000777_050 |
0.0810881981 |
- |
- |
hypothetical protein CICLE_v10008422mg [Citrus clementina] |