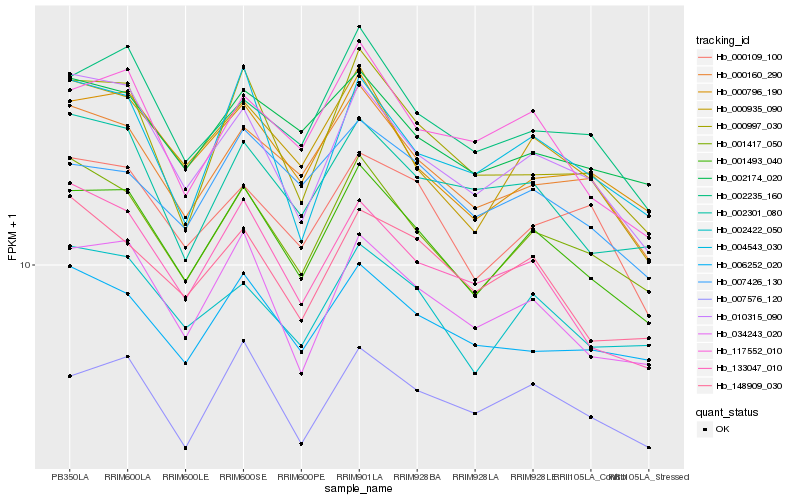
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001493_040 |
0.0 |
- |
- |
PREDICTED: FACT complex subunit SPT16-like [Jatropha curcas] |
| 2 |
Hb_001417_050 |
0.0511002722 |
- |
- |
PREDICTED: G patch domain-containing protein TGH [Jatropha curcas] |
| 3 |
Hb_000935_090 |
0.0602125326 |
- |
- |
PREDICTED: putative nuclear matrix constituent protein 1-like protein [Jatropha curcas] |
| 4 |
Hb_034243_020 |
0.060233753 |
- |
- |
PREDICTED: elongator complex protein 1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_010315_090 |
0.0603576305 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 6 |
Hb_000160_290 |
0.0621324311 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_002422_050 |
0.0635019647 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 5 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000796_190 |
0.0650193906 |
- |
- |
hypothetical protein F383_31148 [Gossypium arboreum] |
| 9 |
Hb_007576_120 |
0.0672412526 |
- |
- |
NBS-LRR disease resistance protein NBS49 [Dimocarpus longan] |
| 10 |
Hb_117552_010 |
0.0701578292 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_002235_160 |
0.0724232942 |
- |
- |
PREDICTED: SNF1-related protein kinase catalytic subunit alpha KIN10 [Jatropha curcas] |
| 12 |
Hb_002174_020 |
0.0729283405 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 13 |
Hb_000997_030 |
0.0742776212 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 14 |
Hb_006252_020 |
0.0751223614 |
- |
- |
PREDICTED: SET and MYND domain-containing protein 4 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000109_100 |
0.0760874793 |
- |
- |
PREDICTED: poly [ADP-ribose] polymerase 1 [Jatropha curcas] |
| 16 |
Hb_004543_030 |
0.076733524 |
- |
- |
PREDICTED: FIP1[V]-like protein [Jatropha curcas] |
| 17 |
Hb_002301_080 |
0.0768601268 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105649589 isoform X2 [Jatropha curcas] |
| 18 |
Hb_148909_030 |
0.0775260811 |
- |
- |
PREDICTED: uncharacterized protein LOC105638192 isoform X1 [Jatropha curcas] |
| 19 |
Hb_133047_010 |
0.0781648659 |
- |
- |
xanthine dehydrogenase, putative [Ricinus communis] |
| 20 |
Hb_007426_130 |
0.0787256537 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |