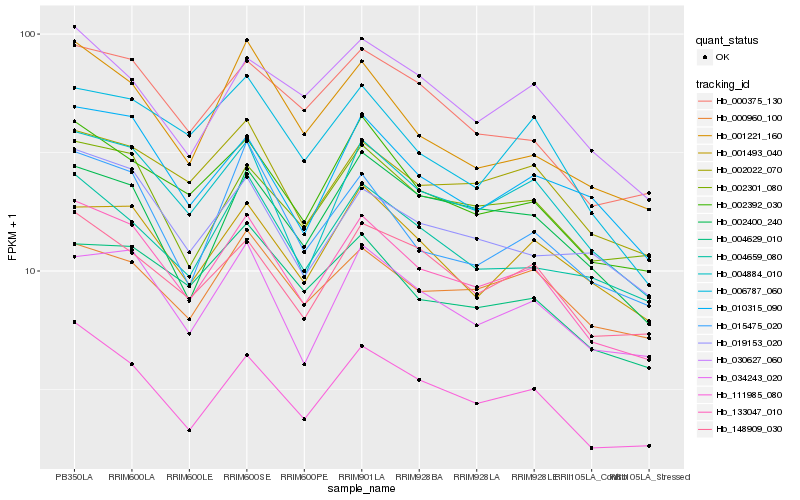
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_133047_010 |
0.0 |
- |
- |
xanthine dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_111985_080 |
0.0513807095 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g19720 [Jatropha curcas] |
| 3 |
Hb_004884_010 |
0.0546383231 |
- |
- |
PREDICTED: pre-mRNA-splicing factor CWC22 homolog [Jatropha curcas] |
| 4 |
Hb_002392_030 |
0.0570924629 |
- |
- |
WD-40 repeat family protein / small nuclear ribonucleoprotein Prp4p-related [Theobroma cacao] |
| 5 |
Hb_034243_020 |
0.068154717 |
- |
- |
PREDICTED: elongator complex protein 1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002301_080 |
0.0684534186 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105649589 isoform X2 [Jatropha curcas] |
| 7 |
Hb_148909_030 |
0.0692228499 |
- |
- |
PREDICTED: uncharacterized protein LOC105638192 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000375_130 |
0.0720312376 |
- |
- |
Leucine-rich repeat transmembrane protein kinase family protein isoform 1 [Theobroma cacao] |
| 9 |
Hb_010315_090 |
0.072394724 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 10 |
Hb_002022_070 |
0.0734906766 |
- |
- |
PREDICTED: histidine--tRNA ligase [Jatropha curcas] |
| 11 |
Hb_015475_020 |
0.0748555819 |
- |
- |
importin beta-1, putative [Ricinus communis] |
| 12 |
Hb_004629_010 |
0.0753712617 |
- |
- |
PREDICTED: AP-3 complex subunit delta [Jatropha curcas] |
| 13 |
Hb_001221_160 |
0.0756443894 |
- |
- |
PREDICTED: exocyst complex component EXO70B1 [Jatropha curcas] |
| 14 |
Hb_004659_080 |
0.0778194001 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3D isoform X1 [Jatropha curcas] |
| 15 |
Hb_019153_020 |
0.0779315261 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 16 |
Hb_001493_040 |
0.0781648659 |
- |
- |
PREDICTED: FACT complex subunit SPT16-like [Jatropha curcas] |
| 17 |
Hb_006787_060 |
0.0798494615 |
- |
- |
PREDICTED: probable E3 ubiquitin ligase SUD1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_030627_060 |
0.0820004054 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 19 |
Hb_000960_100 |
0.0833269248 |
- |
- |
PREDICTED: uncharacterized protein At1g51745 [Jatropha curcas] |
| 20 |
Hb_002400_240 |
0.0838087569 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |