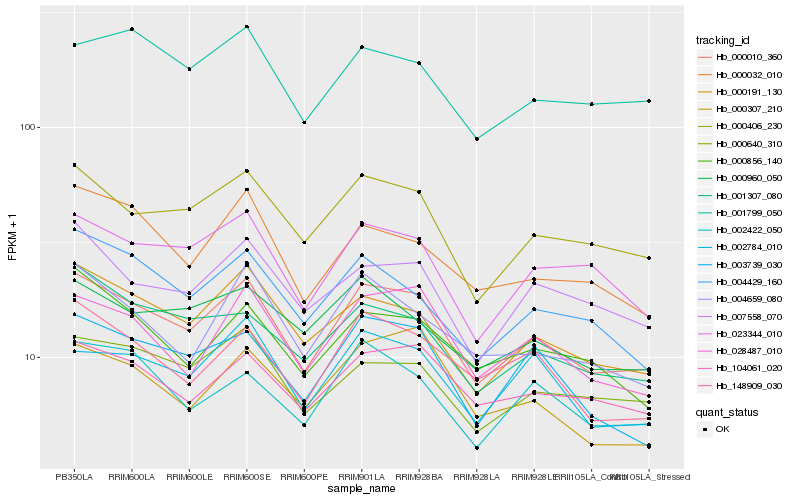
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000010_360 |
0.0 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY [Jatropha curcas] |
| 2 |
Hb_000406_230 |
0.0543476815 |
- |
- |
PREDICTED: probable nucleolar protein 5-1 [Jatropha curcas] |
| 3 |
Hb_000191_130 |
0.0560400341 |
- |
- |
PREDICTED: RNA polymerase-associated protein CTR9 homolog [Jatropha curcas] |
| 4 |
Hb_028487_010 |
0.0648456493 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 23 isoform X2 [Jatropha curcas] |
| 5 |
Hb_023344_010 |
0.065163122 |
- |
- |
PREDICTED: pre-mRNA-processing factor 39 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000640_310 |
0.0677340783 |
- |
- |
hypothetical protein JCGZ_14055 [Jatropha curcas] |
| 7 |
Hb_104061_020 |
0.0695794026 |
- |
- |
PREDICTED: UV-stimulated scaffold protein A homolog [Jatropha curcas] |
| 8 |
Hb_148909_030 |
0.0698704429 |
- |
- |
PREDICTED: uncharacterized protein LOC105638192 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003739_030 |
0.0698919266 |
- |
- |
PREDICTED: putative pre-mRNA-splicing factor ATP-dependent RNA helicase DHX16 [Jatropha curcas] |
| 10 |
Hb_007558_070 |
0.0699212205 |
- |
- |
PREDICTED: pre-mRNA-processing factor 39 isoform X2 [Jatropha curcas] |
| 11 |
Hb_002784_010 |
0.0709770777 |
- |
- |
DNA-directed RNA polymerase I subunit, putative [Ricinus communis] |
| 12 |
Hb_001307_080 |
0.0724211361 |
- |
- |
PREDICTED: phospholipid--sterol O-acyltransferase [Jatropha curcas] |
| 13 |
Hb_002422_050 |
0.0737874141 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 5 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000032_010 |
0.0740405917 |
- |
- |
crooked neck protein, putative [Ricinus communis] |
| 15 |
Hb_004429_160 |
0.0743233223 |
- |
- |
PREDICTED: serine/threonine-protein kinase TAO3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001799_050 |
0.0743280725 |
- |
- |
PREDICTED: plasminogen activator inhibitor 1 RNA-binding protein [Vitis vinifera] |
| 17 |
Hb_000960_050 |
0.0751070138 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |
| 18 |
Hb_000307_210 |
0.0761700282 |
- |
- |
Transportin 1 isoform 1 [Theobroma cacao] |
| 19 |
Hb_000856_140 |
0.0773062146 |
- |
- |
PREDICTED: uncharacterized protein LOC105640478 isoform X1 [Jatropha curcas] |
| 20 |
Hb_004659_080 |
0.0773834432 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3D isoform X1 [Jatropha curcas] |