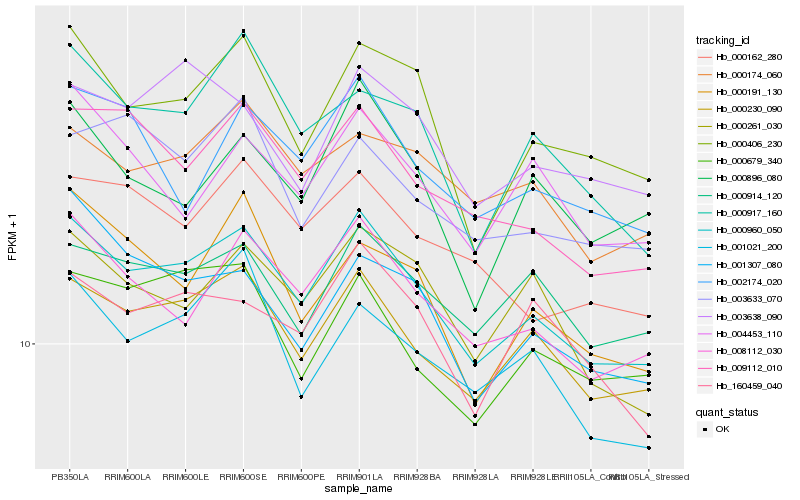
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000960_050 |
0.0 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |
| 2 |
Hb_000230_090 |
0.0410232246 |
- |
- |
PREDICTED: WD repeat-containing protein 48 isoform X1 [Jatropha curcas] |
| 3 |
Hb_008112_030 |
0.0527851351 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_009112_010 |
0.0605599622 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 2 homolog A [Jatropha curcas] |
| 5 |
Hb_000406_230 |
0.060728959 |
- |
- |
PREDICTED: probable nucleolar protein 5-1 [Jatropha curcas] |
| 6 |
Hb_000914_120 |
0.0649697382 |
- |
- |
PREDICTED: outer envelope protein 80, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000174_060 |
0.0662240316 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 8 |
Hb_000261_030 |
0.0666364896 |
- |
- |
PREDICTED: uncharacterized protein LOC105633147 [Jatropha curcas] |
| 9 |
Hb_000917_160 |
0.0689872943 |
- |
- |
PREDICTED: protein decapping 5 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000162_280 |
0.0690277031 |
- |
- |
PREDICTED: CTL-like protein DDB_G0288717 [Populus euphratica] |
| 11 |
Hb_000679_340 |
0.0696431521 |
- |
- |
PREDICTED: uncharacterized protein LOC105650070 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001021_200 |
0.070124231 |
desease resistance |
Gene Name: AAA |
PREDICTED: peroxisome biogenesis protein 6 [Jatropha curcas] |
| 13 |
Hb_160459_040 |
0.0702400783 |
- |
- |
hypothetical protein JCGZ_01511 [Jatropha curcas] |
| 14 |
Hb_000896_080 |
0.0719822829 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP95 isoform X1 [Vitis vinifera] |
| 15 |
Hb_000191_130 |
0.0719949752 |
- |
- |
PREDICTED: RNA polymerase-associated protein CTR9 homolog [Jatropha curcas] |
| 16 |
Hb_004453_110 |
0.0722423649 |
- |
- |
Peptidyl-prolyl cis-trans isomerase CYP19-2 isoform 1 [Theobroma cacao] |
| 17 |
Hb_002174_020 |
0.0723021632 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 18 |
Hb_003638_090 |
0.0725615987 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR45a-like [Jatropha curcas] |
| 19 |
Hb_001307_080 |
0.0738516767 |
- |
- |
PREDICTED: phospholipid--sterol O-acyltransferase [Jatropha curcas] |
| 20 |
Hb_003633_070 |
0.0739440069 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |