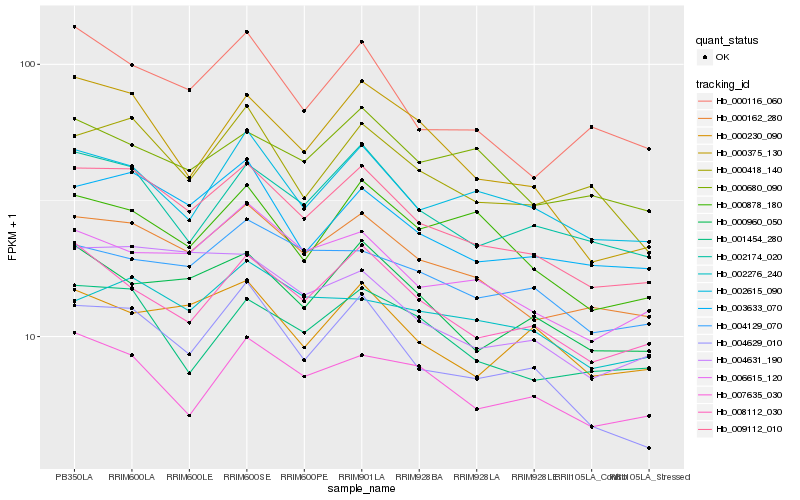
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009112_010 |
0.0 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 2 homolog A [Jatropha curcas] |
| 2 |
Hb_000162_280 |
0.0368357918 |
- |
- |
PREDICTED: CTL-like protein DDB_G0288717 [Populus euphratica] |
| 3 |
Hb_003633_070 |
0.0549359492 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |
| 4 |
Hb_004629_010 |
0.0551462667 |
- |
- |
PREDICTED: AP-3 complex subunit delta [Jatropha curcas] |
| 5 |
Hb_000960_050 |
0.0605599622 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |
| 6 |
Hb_008112_030 |
0.0607395423 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_006615_120 |
0.0622073742 |
- |
- |
PREDICTED: uncharacterized protein LOC105640682 [Jatropha curcas] |
| 8 |
Hb_002174_020 |
0.063161989 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 9 |
Hb_000418_140 |
0.0661180366 |
- |
- |
PREDICTED: DNA damage-binding protein 1a [Jatropha curcas] |
| 10 |
Hb_002615_090 |
0.0674746017 |
- |
- |
PREDICTED: protein SMG7 isoform X1 [Jatropha curcas] |
| 11 |
Hb_004631_190 |
0.0684909499 |
- |
- |
PREDICTED: protein SCAI homolog [Jatropha curcas] |
| 12 |
Hb_001454_280 |
0.0707202725 |
- |
- |
PREDICTED: chaperone protein dnaJ 49 [Jatropha curcas] |
| 13 |
Hb_000116_060 |
0.0709277726 |
- |
- |
plant ubiquilin, putative [Ricinus communis] |
| 14 |
Hb_004129_070 |
0.0718910225 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 5 [Jatropha curcas] |
| 15 |
Hb_000878_180 |
0.0727529111 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 12 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000375_130 |
0.0729945325 |
- |
- |
Leucine-rich repeat transmembrane protein kinase family protein isoform 1 [Theobroma cacao] |
| 17 |
Hb_007635_030 |
0.073363488 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g77360, mitochondrial-like [Jatropha curcas] |
| 18 |
Hb_000230_090 |
0.0734332159 |
- |
- |
PREDICTED: WD repeat-containing protein 48 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002276_240 |
0.0766902259 |
- |
- |
PREDICTED: uncharacterized protein LOC105640682 [Jatropha curcas] |
| 20 |
Hb_000680_090 |
0.0776131991 |
- |
- |
Protein SEY1, putative [Ricinus communis] |