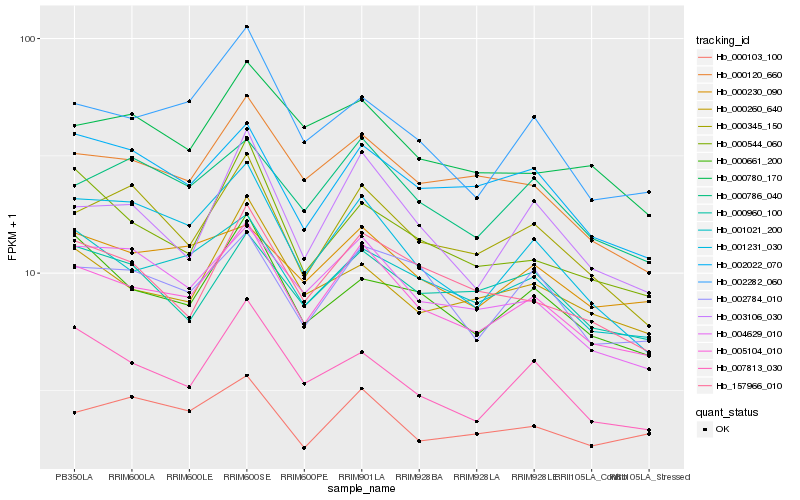
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005104_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105140796 isoform X1 [Populus euphratica] |
| 2 |
Hb_001021_200 |
0.0676774671 |
desease resistance |
Gene Name: AAA |
PREDICTED: peroxisome biogenesis protein 6 [Jatropha curcas] |
| 3 |
Hb_004629_010 |
0.0707820118 |
- |
- |
PREDICTED: AP-3 complex subunit delta [Jatropha curcas] |
| 4 |
Hb_000120_660 |
0.0710042197 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 5 |
Hb_000786_040 |
0.0734881552 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 17 [Jatropha curcas] |
| 6 |
Hb_000345_150 |
0.0737663202 |
- |
- |
PREDICTED: uncharacterized protein LOC105647517 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003106_030 |
0.0743533577 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0271670 [Jatropha curcas] |
| 8 |
Hb_002784_010 |
0.0760829324 |
- |
- |
DNA-directed RNA polymerase I subunit, putative [Ricinus communis] |
| 9 |
Hb_157966_010 |
0.0764007485 |
- |
- |
hypothetical protein JCGZ_13877 [Jatropha curcas] |
| 10 |
Hb_001231_030 |
0.0779067054 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like [Jatropha curcas] |
| 11 |
Hb_002282_060 |
0.0784844557 |
transcription factor |
TF Family: bZIP |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_002022_070 |
0.0794711439 |
- |
- |
PREDICTED: histidine--tRNA ligase [Jatropha curcas] |
| 13 |
Hb_000230_090 |
0.081064492 |
- |
- |
PREDICTED: WD repeat-containing protein 48 isoform X1 [Jatropha curcas] |
| 14 |
Hb_007813_030 |
0.0819639133 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein At4g19050 [Jatropha curcas] |
| 15 |
Hb_000661_200 |
0.0824119244 |
- |
- |
PREDICTED: uncharacterized protein LOC105648311 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000260_640 |
0.0824668146 |
- |
- |
PREDICTED: uncharacterized protein At1g51745 [Jatropha curcas] |
| 17 |
Hb_000960_100 |
0.0845557957 |
- |
- |
PREDICTED: uncharacterized protein At1g51745 [Jatropha curcas] |
| 18 |
Hb_000103_100 |
0.0851453867 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g22410, mitochondrial-like [Jatropha curcas] |
| 19 |
Hb_000544_060 |
0.085384918 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 20 |
Hb_000780_170 |
0.0856408369 |
- |
- |
PREDICTED: OBERON-like protein [Jatropha curcas] |