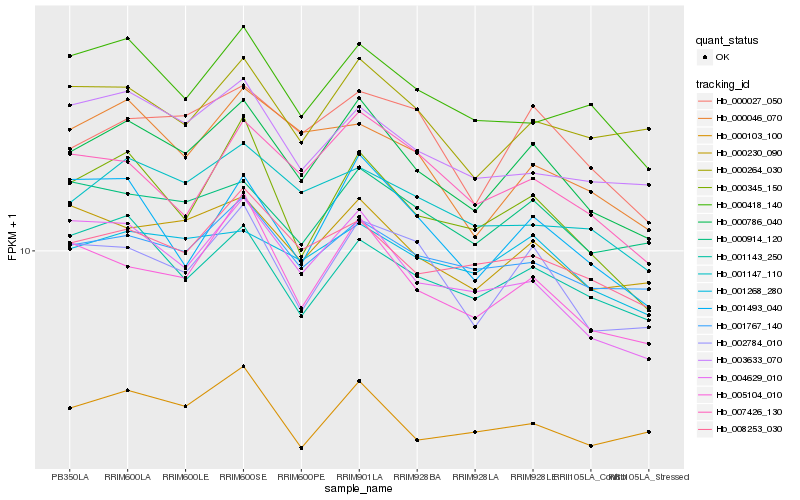
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000786_040 |
0.0 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 17 [Jatropha curcas] |
| 2 |
Hb_002784_010 |
0.0702029966 |
- |
- |
DNA-directed RNA polymerase I subunit, putative [Ricinus communis] |
| 3 |
Hb_000230_090 |
0.0710117362 |
- |
- |
PREDICTED: WD repeat-containing protein 48 isoform X1 [Jatropha curcas] |
| 4 |
Hb_005104_010 |
0.0734881552 |
- |
- |
PREDICTED: uncharacterized protein LOC105140796 isoform X1 [Populus euphratica] |
| 5 |
Hb_008253_030 |
0.0750134794 |
- |
- |
PREDICTED: probable carboxylesterase 11 [Jatropha curcas] |
| 6 |
Hb_001767_140 |
0.0751350456 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |
| 7 |
Hb_000345_150 |
0.0751527072 |
- |
- |
PREDICTED: uncharacterized protein LOC105647517 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000027_050 |
0.0763250113 |
- |
- |
PREDICTED: uncharacterized protein LOC105636933 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001268_280 |
0.0763818494 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_004629_010 |
0.0783190294 |
- |
- |
PREDICTED: AP-3 complex subunit delta [Jatropha curcas] |
| 11 |
Hb_000914_120 |
0.0793680123 |
- |
- |
PREDICTED: outer envelope protein 80, chloroplastic [Jatropha curcas] |
| 12 |
Hb_001147_110 |
0.0798576814 |
- |
- |
PREDICTED: dynamin-2A-like [Jatropha curcas] |
| 13 |
Hb_001143_250 |
0.0809800387 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3C isoform X1 [Jatropha curcas] |
| 14 |
Hb_001493_040 |
0.0825559716 |
- |
- |
PREDICTED: FACT complex subunit SPT16-like [Jatropha curcas] |
| 15 |
Hb_000418_140 |
0.0827464601 |
- |
- |
PREDICTED: DNA damage-binding protein 1a [Jatropha curcas] |
| 16 |
Hb_000264_030 |
0.0827646345 |
- |
- |
PREDICTED: splicing factor 3B subunit 2 [Jatropha curcas] |
| 17 |
Hb_000103_100 |
0.083111219 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g22410, mitochondrial-like [Jatropha curcas] |
| 18 |
Hb_003633_070 |
0.0842807954 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |
| 19 |
Hb_007426_130 |
0.0844841109 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 20 |
Hb_000046_070 |
0.0853730396 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |