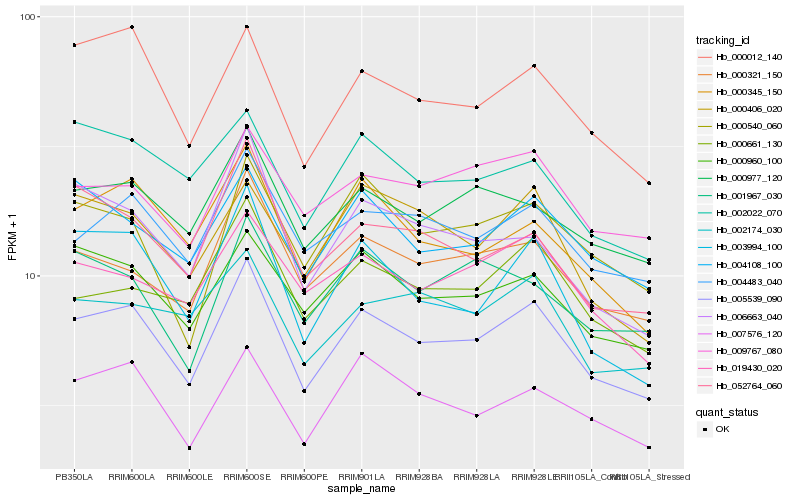
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005539_090 |
0.0 |
transcription factor |
TF Family: PHD |
PREDICTED: methyl-CpG-binding domain-containing protein 9 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000012_140 |
0.0701456132 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_007576_120 |
0.0722222815 |
- |
- |
NBS-LRR disease resistance protein NBS49 [Dimocarpus longan] |
| 4 |
Hb_004483_040 |
0.0722986397 |
- |
- |
hypothetical protein JCGZ_22047 [Jatropha curcas] |
| 5 |
Hb_000977_120 |
0.074450266 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 4 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000321_150 |
0.0749598983 |
- |
- |
PREDICTED: uncharacterized protein LOC105645596 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000960_100 |
0.0763299175 |
- |
- |
PREDICTED: uncharacterized protein At1g51745 [Jatropha curcas] |
| 8 |
Hb_000345_150 |
0.0770045806 |
- |
- |
PREDICTED: uncharacterized protein LOC105647517 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003994_100 |
0.0770460215 |
transcription factor |
TF Family: PHD |
PREDICTED: histone acetyltransferase HAC1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_004108_100 |
0.0783768395 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33B isoform X2 [Jatropha curcas] |
| 11 |
Hb_000661_130 |
0.0807596952 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105648320 [Jatropha curcas] |
| 12 |
Hb_019430_020 |
0.082843411 |
- |
- |
PREDICTED: uncharacterized protein LOC105641844 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000540_060 |
0.0837396011 |
- |
- |
PREDICTED: uncharacterized protein LOC105628177 [Jatropha curcas] |
| 14 |
Hb_009767_080 |
0.0837887061 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631983 [Jatropha curcas] |
| 15 |
Hb_002174_030 |
0.0859836882 |
- |
- |
transcription elongation factor s-II, putative [Ricinus communis] |
| 16 |
Hb_052764_060 |
0.086226161 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 17 |
Hb_002022_070 |
0.0867058073 |
- |
- |
PREDICTED: histidine--tRNA ligase [Jatropha curcas] |
| 18 |
Hb_000406_020 |
0.0873959777 |
- |
- |
- |
| 19 |
Hb_006663_040 |
0.0892041027 |
- |
- |
PREDICTED: DDB1- and CUL4-associated factor homolog 1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001967_030 |
0.08962469 |
- |
- |
PREDICTED: uncharacterized protein LOC105633557 [Jatropha curcas] |