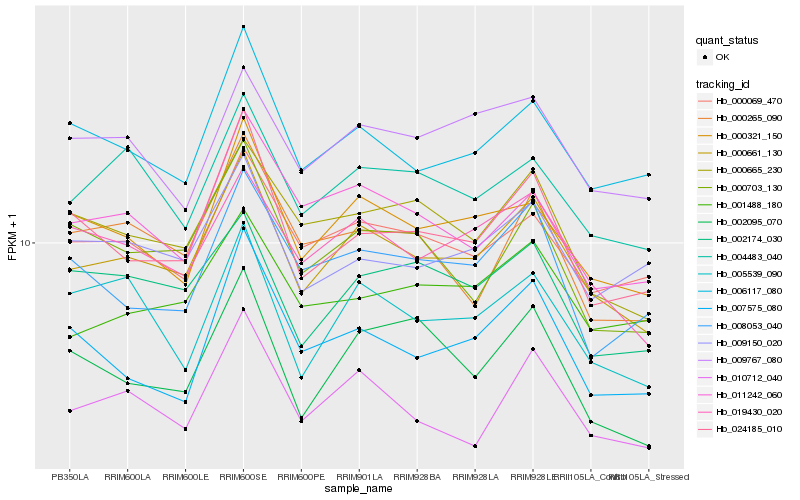
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000661_130 |
0.0 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105648320 [Jatropha curcas] |
| 2 |
Hb_019430_020 |
0.0637655632 |
- |
- |
PREDICTED: uncharacterized protein LOC105641844 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000321_150 |
0.0647030997 |
- |
- |
PREDICTED: uncharacterized protein LOC105645596 isoform X1 [Jatropha curcas] |
| 4 |
Hb_006117_080 |
0.0712323471 |
- |
- |
PREDICTED: transcription factor GTE4 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000069_470 |
0.0714911368 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 6 |
Hb_010712_040 |
0.0715540174 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g11290 [Jatropha curcas] |
| 7 |
Hb_004483_040 |
0.0719213668 |
- |
- |
hypothetical protein JCGZ_22047 [Jatropha curcas] |
| 8 |
Hb_024185_010 |
0.0725996782 |
transcription factor |
TF Family: PHD |
PREDICTED: histone acetyltransferase HAC1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000665_230 |
0.0728650344 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 10 |
Hb_002174_030 |
0.0786355935 |
- |
- |
transcription elongation factor s-II, putative [Ricinus communis] |
| 11 |
Hb_011242_060 |
0.0797589952 |
transcription factor |
TF Family: SNF2 |
Chromo domain protein, putative [Ricinus communis] |
| 12 |
Hb_001488_180 |
0.0797761255 |
- |
- |
PREDICTED: uncharacterized protein LOC105644152 isoform X1 [Jatropha curcas] |
| 13 |
Hb_009767_080 |
0.0800931893 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631983 [Jatropha curcas] |
| 14 |
Hb_005539_090 |
0.0807596952 |
transcription factor |
TF Family: PHD |
PREDICTED: methyl-CpG-binding domain-containing protein 9 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000703_130 |
0.0820937817 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DHX36 [Jatropha curcas] |
| 16 |
Hb_009150_020 |
0.082396385 |
- |
- |
PREDICTED: uncharacterized protein LOC105630583 [Jatropha curcas] |
| 17 |
Hb_008053_040 |
0.0850537885 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 12-like [Pyrus x bretschneideri] |
| 18 |
Hb_000265_090 |
0.0860032311 |
- |
- |
PREDICTED: flowering time control protein FPA [Jatropha curcas] |
| 19 |
Hb_007575_080 |
0.086956999 |
- |
- |
PREDICTED: lysine-specific histone demethylase 1 homolog 3 [Jatropha curcas] |
| 20 |
Hb_002095_070 |
0.0889600042 |
- |
- |
PREDICTED: probable pre-mRNA-splicing factor ATP-dependent RNA helicase [Jatropha curcas] |