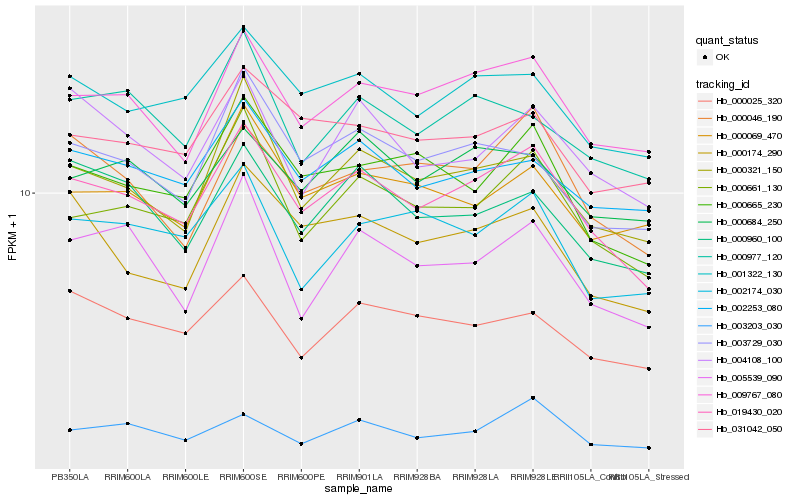
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_019430_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641844 isoform X1 [Jatropha curcas] |
| 2 |
Hb_009767_080 |
0.0604980206 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631983 [Jatropha curcas] |
| 3 |
Hb_000661_130 |
0.0637655632 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105648320 [Jatropha curcas] |
| 4 |
Hb_001322_130 |
0.0657180697 |
- |
- |
PREDICTED: putative glycerol-3-phosphate transporter 4 [Jatropha curcas] |
| 5 |
Hb_000665_230 |
0.0679730588 |
- |
- |
PREDICTED: uncharacterized protein LOC105637615 [Jatropha curcas] |
| 6 |
Hb_000321_150 |
0.0712588086 |
- |
- |
PREDICTED: uncharacterized protein LOC105645596 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002253_080 |
0.0749785177 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase beta 1-like [Jatropha curcas] |
| 8 |
Hb_000046_190 |
0.0761223071 |
- |
- |
PREDICTED: uncharacterized protein LOC105647118 [Jatropha curcas] |
| 9 |
Hb_003729_030 |
0.0768852451 |
- |
- |
PREDICTED: alpha,alpha-trehalose-phosphate synthase [UDP-forming] 6 [Jatropha curcas] |
| 10 |
Hb_000174_290 |
0.078896677 |
- |
- |
PREDICTED: conserved oligomeric Golgi complex subunit 1 [Jatropha curcas] |
| 11 |
Hb_031042_050 |
0.0795026026 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 12 |
Hb_004108_100 |
0.0797727481 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33B isoform X2 [Jatropha curcas] |
| 13 |
Hb_000684_250 |
0.0800155229 |
- |
- |
PREDICTED: uncharacterized protein LOC105642172 [Jatropha curcas] |
| 14 |
Hb_003203_030 |
0.0807393748 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g13600-like [Jatropha curcas] |
| 15 |
Hb_000025_320 |
0.081111897 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g28010 [Jatropha curcas] |
| 16 |
Hb_000960_100 |
0.0814472587 |
- |
- |
PREDICTED: uncharacterized protein At1g51745 [Jatropha curcas] |
| 17 |
Hb_000977_120 |
0.0818346518 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 4 isoform X2 [Jatropha curcas] |
| 18 |
Hb_005539_090 |
0.082843411 |
transcription factor |
TF Family: PHD |
PREDICTED: methyl-CpG-binding domain-containing protein 9 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002174_030 |
0.0833121558 |
- |
- |
transcription elongation factor s-II, putative [Ricinus communis] |
| 20 |
Hb_000069_470 |
0.083366707 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |