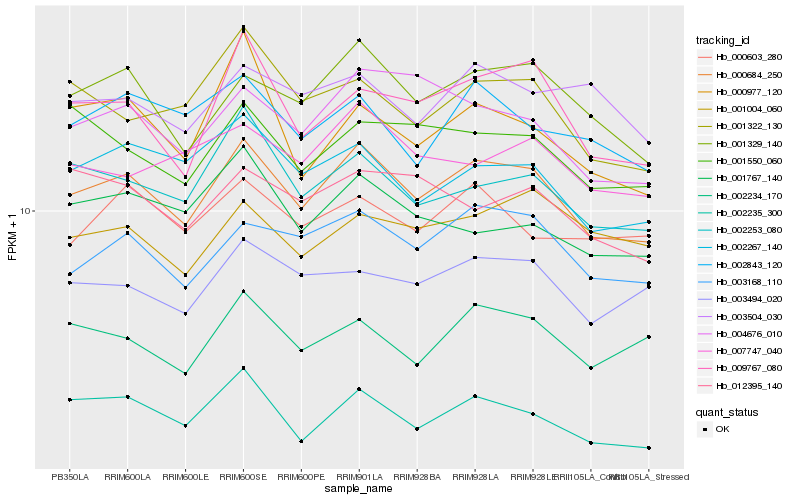
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000684_250 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642172 [Jatropha curcas] |
| 2 |
Hb_001329_140 |
0.0531227795 |
- |
- |
PREDICTED: zinc finger protein 598 [Jatropha curcas] |
| 3 |
Hb_003168_110 |
0.0564118499 |
transcription factor |
TF Family: SWI/SNF-SWI3 |
PREDICTED: lysine-specific histone demethylase 1 homolog 1 [Jatropha curcas] |
| 4 |
Hb_002235_300 |
0.057025236 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15280 isoform X1 [Jatropha curcas] |
| 5 |
Hb_009767_080 |
0.0630588875 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631983 [Jatropha curcas] |
| 6 |
Hb_002253_080 |
0.0644973888 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase beta 1-like [Jatropha curcas] |
| 7 |
Hb_002267_140 |
0.0656834397 |
- |
- |
PREDICTED: VHS domain-containing protein At3g16270 [Jatropha curcas] |
| 8 |
Hb_004676_010 |
0.0682430832 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g80270, mitochondrial-like [Jatropha curcas] |
| 9 |
Hb_003494_020 |
0.0685257133 |
- |
- |
ATP-dependent RNA and DNA helicase, putative [Ricinus communis] |
| 10 |
Hb_002234_170 |
0.0700882374 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_002843_120 |
0.0712905019 |
- |
- |
PREDICTED: uncharacterized protein LOC105645113 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001322_130 |
0.0713511845 |
- |
- |
PREDICTED: putative glycerol-3-phosphate transporter 4 [Jatropha curcas] |
| 13 |
Hb_001550_060 |
0.0723931347 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL3 isoform X4 [Jatropha curcas] |
| 14 |
Hb_007747_040 |
0.0744598229 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY [Jatropha curcas] |
| 15 |
Hb_000603_280 |
0.0753172483 |
- |
- |
PREDICTED: uncharacterized protein LOC105638082 [Jatropha curcas] |
| 16 |
Hb_001767_140 |
0.0762843435 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |
| 17 |
Hb_012395_140 |
0.0767338554 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |
| 18 |
Hb_001004_060 |
0.0770741023 |
- |
- |
PREDICTED: uncharacterized protein LOC105630220 isoform X4 [Jatropha curcas] |
| 19 |
Hb_003504_030 |
0.0778301085 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_000977_120 |
0.0779793175 |
- |
- |
PREDICTED: sister chromatid cohesion 1 protein 4 isoform X2 [Jatropha curcas] |