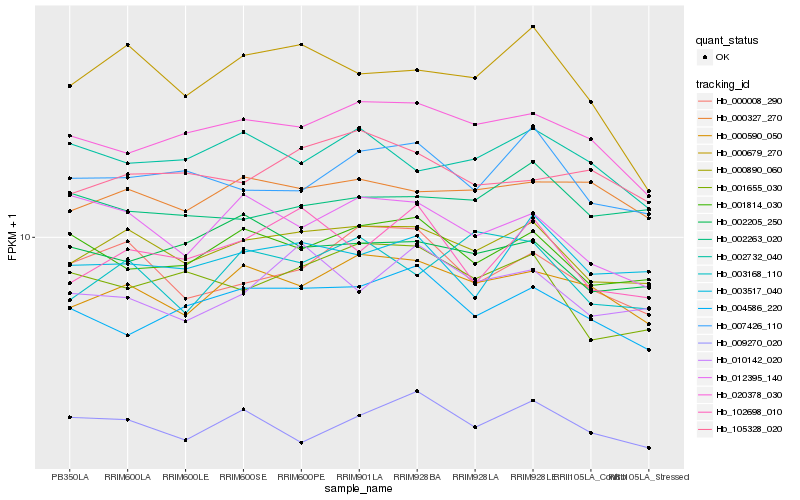
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000890_060 |
0.0 |
- |
- |
PREDICTED: sucrose nonfermenting 4-like protein [Jatropha curcas] |
| 2 |
Hb_000590_050 |
0.0517917006 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |
| 3 |
Hb_000008_290 |
0.0579481215 |
- |
- |
PREDICTED: protein SAND [Jatropha curcas] |
| 4 |
Hb_000327_270 |
0.059567675 |
- |
- |
PREDICTED: leukocyte receptor cluster member 8 homolog isoform X1 [Jatropha curcas] |
| 5 |
Hb_001814_030 |
0.0602705978 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |
| 6 |
Hb_007426_110 |
0.0604054211 |
- |
- |
PREDICTED: vacuolar fusion protein CCZ1 homolog isoform X2 [Jatropha curcas] |
| 7 |
Hb_020378_030 |
0.0606888194 |
- |
- |
PREDICTED: protein transport protein SEC31 homolog B [Jatropha curcas] |
| 8 |
Hb_003517_040 |
0.060955015 |
- |
- |
PREDICTED: putative DUF21 domain-containing protein At3g13070, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001655_030 |
0.0630361589 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 3 isoform X2 [Jatropha curcas] |
| 10 |
Hb_002205_250 |
0.0633485594 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase scy1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_102698_010 |
0.065255834 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 5-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_105328_020 |
0.0665627066 |
- |
- |
PREDICTED: phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN [Jatropha curcas] |
| 13 |
Hb_004586_220 |
0.066973976 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |
| 14 |
Hb_012395_140 |
0.067734035 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |
| 15 |
Hb_003168_110 |
0.0683869231 |
transcription factor |
TF Family: SWI/SNF-SWI3 |
PREDICTED: lysine-specific histone demethylase 1 homolog 1 [Jatropha curcas] |
| 16 |
Hb_002263_020 |
0.0684138515 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |
| 17 |
Hb_009270_020 |
0.0684833585 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: helicase and polymerase-containing protein TEBICHI [Jatropha curcas] |
| 18 |
Hb_010142_020 |
0.0690847881 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002732_040 |
0.0705421905 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000679_270 |
0.0715614483 |
- |
- |
PREDICTED: putative clathrin assembly protein At2g25430 [Jatropha curcas] |