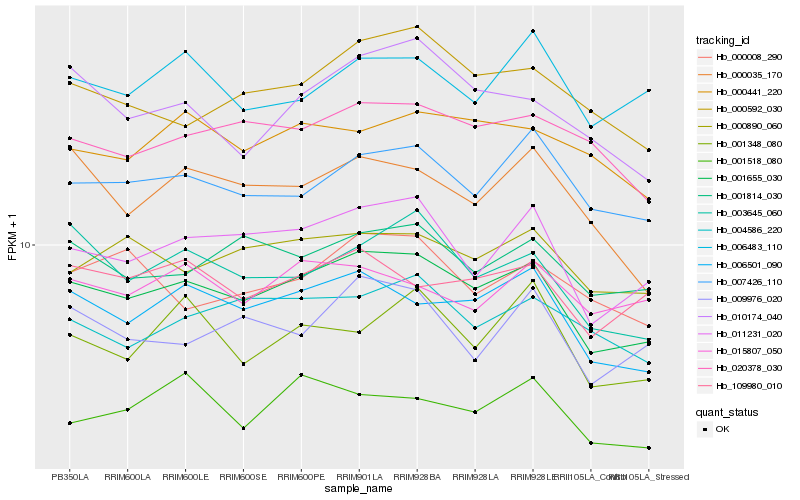
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001655_030 |
0.0 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 3 isoform X2 [Jatropha curcas] |
| 2 |
Hb_006501_090 |
0.0468593786 |
- |
- |
PREDICTED: golgin candidate 1 [Jatropha curcas] |
| 3 |
Hb_011231_020 |
0.0484872191 |
- |
- |
PREDICTED: proline-, glutamic acid- and leucine-rich protein 1 isoform X2 [Jatropha curcas] |
| 4 |
Hb_007426_110 |
0.0595083642 |
- |
- |
PREDICTED: vacuolar fusion protein CCZ1 homolog isoform X2 [Jatropha curcas] |
| 5 |
Hb_001814_030 |
0.0616059134 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |
| 6 |
Hb_003645_060 |
0.0616722999 |
- |
- |
PREDICTED: monodehydroascorbate reductase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000890_060 |
0.0630361589 |
- |
- |
PREDICTED: sucrose nonfermenting 4-like protein [Jatropha curcas] |
| 8 |
Hb_020378_030 |
0.0647710618 |
- |
- |
PREDICTED: protein transport protein SEC31 homolog B [Jatropha curcas] |
| 9 |
Hb_109980_010 |
0.0648724338 |
- |
- |
PREDICTED: uncharacterized protein LOC105647182 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000592_030 |
0.0660802407 |
- |
- |
PREDICTED: uncharacterized protein LOC100853969 isoform X1 [Vitis vinifera] |
| 11 |
Hb_004586_220 |
0.0668451247 |
- |
- |
PREDICTED: WD and tetratricopeptide repeats protein 1 isoform X3 [Jatropha curcas] |
| 12 |
Hb_010174_040 |
0.0685670406 |
- |
- |
PREDICTED: T-complex protein 1 subunit epsilon [Jatropha curcas] |
| 13 |
Hb_000441_220 |
0.0691867104 |
- |
- |
PREDICTED: histidinol dehydrogenase, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_009976_020 |
0.0695836658 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105631286 [Jatropha curcas] |
| 15 |
Hb_000035_170 |
0.0702801281 |
- |
- |
PREDICTED: DNA-directed RNA polymerases IV and V subunit 2-like [Jatropha curcas] |
| 16 |
Hb_001518_080 |
0.0712308044 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646545 [Jatropha curcas] |
| 17 |
Hb_006483_110 |
0.0728854512 |
- |
- |
PREDICTED: RNA-binding protein Musashi homolog 1 [Jatropha curcas] |
| 18 |
Hb_015807_050 |
0.0731519291 |
- |
- |
PREDICTED: uncharacterized protein LOC105632878 [Jatropha curcas] |
| 19 |
Hb_001348_080 |
0.073454001 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 20 |
Hb_000008_290 |
0.0735924278 |
- |
- |
PREDICTED: protein SAND [Jatropha curcas] |