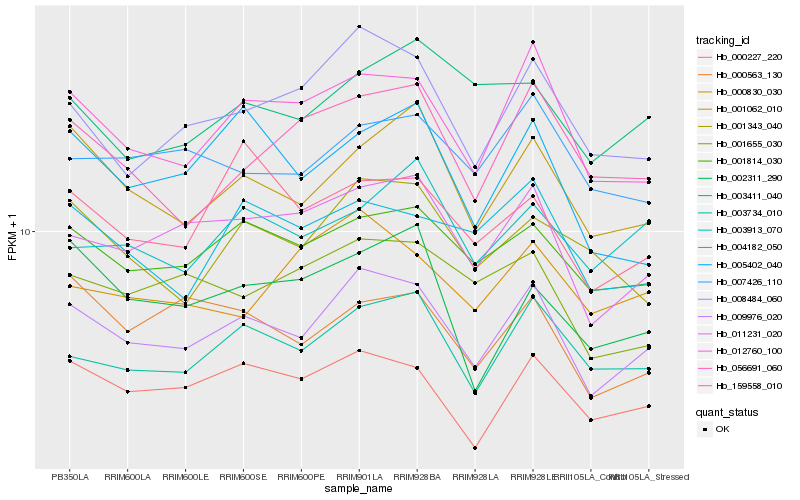
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009976_020 |
0.0 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105631286 [Jatropha curcas] |
| 2 |
Hb_011231_020 |
0.0534874735 |
- |
- |
PREDICTED: proline-, glutamic acid- and leucine-rich protein 1 isoform X2 [Jatropha curcas] |
| 3 |
Hb_012760_100 |
0.0536016518 |
- |
- |
Pre-rRNA-processing protein ESF1, putative [Ricinus communis] |
| 4 |
Hb_001814_030 |
0.0549219171 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |
| 5 |
Hb_000227_220 |
0.0559397827 |
- |
- |
PREDICTED: protein phosphatase 2C 70 [Jatropha curcas] |
| 6 |
Hb_003734_010 |
0.0567953495 |
- |
- |
PREDICTED: double-strand break repair protein MRE11 [Jatropha curcas] |
| 7 |
Hb_001062_010 |
0.0584470478 |
- |
- |
PREDICTED: CWF19-like protein 2 [Jatropha curcas] |
| 8 |
Hb_008484_060 |
0.060314854 |
- |
- |
PREDICTED: allantoate deiminase isoform X3 [Populus euphratica] |
| 9 |
Hb_001655_030 |
0.0695836658 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 3 isoform X2 [Jatropha curcas] |
| 10 |
Hb_007426_110 |
0.0701772507 |
- |
- |
PREDICTED: vacuolar fusion protein CCZ1 homolog isoform X2 [Jatropha curcas] |
| 11 |
Hb_004182_050 |
0.0711645367 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL3 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000563_130 |
0.0720284646 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 13 |
Hb_056691_060 |
0.0727440768 |
- |
- |
PREDICTED: probable manganese-transporting ATPase PDR2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001343_040 |
0.0730835668 |
- |
- |
PREDICTED: uncharacterized protein LOC105638555 [Jatropha curcas] |
| 15 |
Hb_159558_010 |
0.0742947706 |
- |
- |
PREDICTED: uncharacterized protein LOC105635846 [Jatropha curcas] |
| 16 |
Hb_000830_030 |
0.0748398563 |
- |
- |
PREDICTED: uncharacterized protein LOC105644214 [Jatropha curcas] |
| 17 |
Hb_003913_070 |
0.0757142727 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 30 [Populus euphratica] |
| 18 |
Hb_005402_040 |
0.0769525393 |
- |
- |
PREDICTED: replication factor C subunit 1 [Jatropha curcas] |
| 19 |
Hb_002311_290 |
0.0771002695 |
- |
- |
PREDICTED: pumilio homolog 24 [Jatropha curcas] |
| 20 |
Hb_003411_040 |
0.0773654287 |
- |
- |
unnamed protein product [Coffea canephora] |