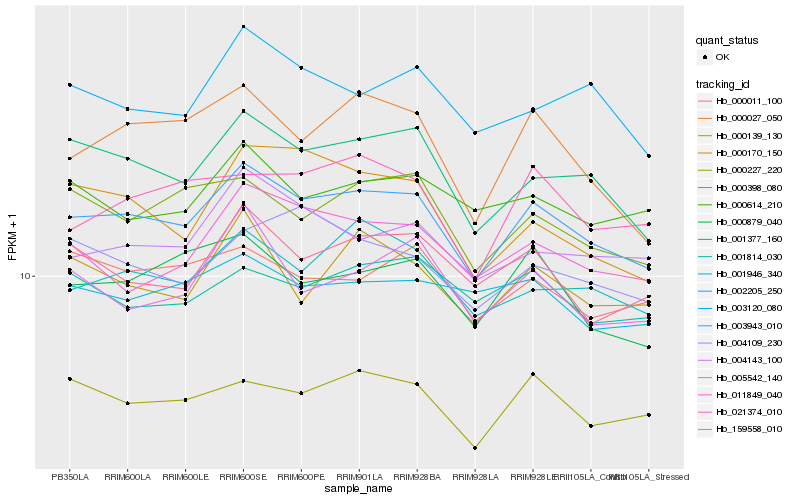
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003943_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001377_160 |
0.0493825634 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 3 |
Hb_001946_340 |
0.0587997867 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 17-like isoform X1 [Jatropha curcas] |
| 4 |
Hb_000227_220 |
0.0603270179 |
- |
- |
PREDICTED: protein phosphatase 2C 70 [Jatropha curcas] |
| 5 |
Hb_000398_080 |
0.0627797563 |
- |
- |
tip120, putative [Ricinus communis] |
| 6 |
Hb_000879_040 |
0.0636280871 |
- |
- |
PREDICTED: calcium-transporting ATPase 3, endoplasmic reticulum-type [Jatropha curcas] |
| 7 |
Hb_159558_010 |
0.0637879275 |
- |
- |
PREDICTED: uncharacterized protein LOC105635846 [Jatropha curcas] |
| 8 |
Hb_011849_040 |
0.064188864 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 9 |
Hb_004143_100 |
0.0648769658 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000027_050 |
0.0654659108 |
- |
- |
PREDICTED: uncharacterized protein LOC105636933 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003120_080 |
0.066844866 |
- |
- |
PREDICTED: COPII coat assembly protein SEC16 isoform X1 [Jatropha curcas] |
| 12 |
Hb_021374_010 |
0.0671217449 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-A1 [Populus euphratica] |
| 13 |
Hb_000170_150 |
0.0678021978 |
- |
- |
PREDICTED: uncharacterized protein LOC105638996 isoform X3 [Jatropha curcas] |
| 14 |
Hb_004109_230 |
0.0680602664 |
- |
- |
heat shock protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_000011_100 |
0.0699658428 |
- |
- |
PREDICTED: uncharacterized protein C630.12 [Jatropha curcas] |
| 16 |
Hb_000614_210 |
0.0705686798 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_005542_140 |
0.0712426486 |
- |
- |
PREDICTED: U-box domain-containing protein 62-like [Jatropha curcas] |
| 18 |
Hb_001814_030 |
0.0719328143 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |
| 19 |
Hb_000139_130 |
0.0719629635 |
- |
- |
PREDICTED: chloride channel protein CLC-d isoform X1 [Jatropha curcas] |
| 20 |
Hb_002205_250 |
0.0719752684 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase scy1 isoform X1 [Jatropha curcas] |