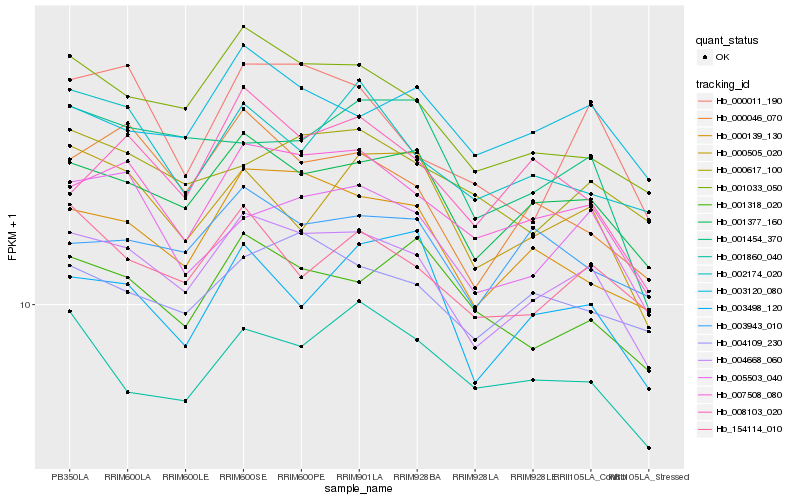
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004668_060 |
0.0 |
- |
- |
PREDICTED: probable apyrase 6 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001377_160 |
0.0506762233 |
- |
- |
PREDICTED: protein SDE2 homolog [Jatropha curcas] |
| 3 |
Hb_007508_080 |
0.0533350754 |
- |
- |
PREDICTED: presenilin-like protein At2g29900 [Jatropha curcas] |
| 4 |
Hb_001033_050 |
0.0642573487 |
- |
- |
PREDICTED: acetylornithine deacetylase [Jatropha curcas] |
| 5 |
Hb_000139_130 |
0.0655915228 |
- |
- |
PREDICTED: chloride channel protein CLC-d isoform X1 [Jatropha curcas] |
| 6 |
Hb_005503_040 |
0.0729962254 |
- |
- |
PREDICTED: AMP deaminase [Jatropha curcas] |
| 7 |
Hb_000046_070 |
0.0764291599 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 8 |
Hb_154114_010 |
0.0783457115 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3A isoform X2 [Jatropha curcas] |
| 9 |
Hb_004109_230 |
0.0789151756 |
- |
- |
heat shock protein binding protein, putative [Ricinus communis] |
| 10 |
Hb_003120_080 |
0.0807312491 |
- |
- |
PREDICTED: COPII coat assembly protein SEC16 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003943_010 |
0.0824865027 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000505_020 |
0.0857772726 |
- |
- |
PREDICTED: protein S-acyltransferase 24 [Jatropha curcas] |
| 13 |
Hb_001318_020 |
0.0859939054 |
- |
- |
HAT dimerization domain-containing protein isoform 2 [Theobroma cacao] |
| 14 |
Hb_003498_120 |
0.0866749461 |
- |
- |
hypothetical protein POPTR_0015s12430g, partial [Populus trichocarpa] |
| 15 |
Hb_000011_190 |
0.0871641894 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 6 [Jatropha curcas] |
| 16 |
Hb_000617_100 |
0.0886169525 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |
| 17 |
Hb_001454_370 |
0.0887515229 |
- |
- |
PREDICTED: glyoxysomal fatty acid beta-oxidation multifunctional protein MFP-a [Jatropha curcas] |
| 18 |
Hb_001860_040 |
0.0896733781 |
- |
- |
PREDICTED: cleavage stimulation factor subunit 77 isoform X4 [Jatropha curcas] |
| 19 |
Hb_002174_020 |
0.0900387252 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 20 |
Hb_008103_020 |
0.0900605394 |
- |
- |
hypothetical protein POPTR_0003s20310g [Populus trichocarpa] |