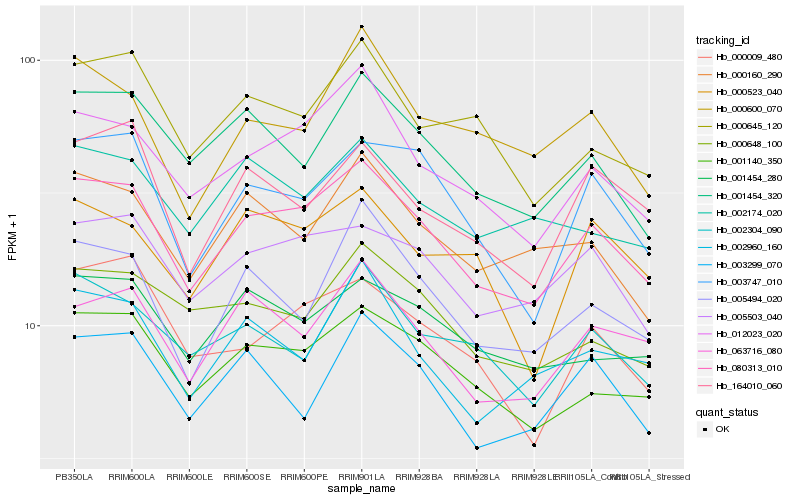
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_080313_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105634976 [Jatropha curcas] |
| 2 |
Hb_012023_020 |
0.0584427086 |
- |
- |
hypothetical protein JCGZ_03076 [Jatropha curcas] |
| 3 |
Hb_001140_350 |
0.0622428669 |
- |
- |
PREDICTED: uncharacterized protein LOC105648465 [Jatropha curcas] |
| 4 |
Hb_005503_040 |
0.0657488978 |
- |
- |
PREDICTED: AMP deaminase [Jatropha curcas] |
| 5 |
Hb_063716_080 |
0.0766608753 |
- |
- |
DNA-directed RNA polymerase III subunit F, putative [Ricinus communis] |
| 6 |
Hb_003747_010 |
0.0791572831 |
- |
- |
hypothetical protein JCGZ_24080 [Jatropha curcas] |
| 7 |
Hb_001454_320 |
0.079437374 |
- |
- |
PREDICTED: protein pleiotropic regulatory locus 1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001454_280 |
0.081400841 |
- |
- |
PREDICTED: chaperone protein dnaJ 49 [Jatropha curcas] |
| 9 |
Hb_005494_020 |
0.0816858932 |
- |
- |
PREDICTED: transmembrane protein 184 homolog DDB_G0279555 [Jatropha curcas] |
| 10 |
Hb_000160_290 |
0.0828089755 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_000648_100 |
0.0847548699 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 12 |
Hb_002960_160 |
0.085262348 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 35B isoform X2 [Jatropha curcas] |
| 13 |
Hb_000645_120 |
0.085298034 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 32 [Jatropha curcas] |
| 14 |
Hb_002304_090 |
0.0860058934 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DHX35 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000009_480 |
0.0870564008 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g66560 [Jatropha curcas] |
| 16 |
Hb_002174_020 |
0.0879497429 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 17 |
Hb_003299_070 |
0.0880655485 |
- |
- |
AMP deaminase [Theobroma cacao] |
| 18 |
Hb_164010_060 |
0.0883370045 |
- |
- |
PREDICTED: uncharacterized protein LOC105633115 [Jatropha curcas] |
| 19 |
Hb_000523_040 |
0.0897421811 |
- |
- |
PREDICTED: auxin response factor 8 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000600_070 |
0.0898013568 |
- |
- |
PREDICTED: leucine aminopeptidase 1-like [Jatropha curcas] |