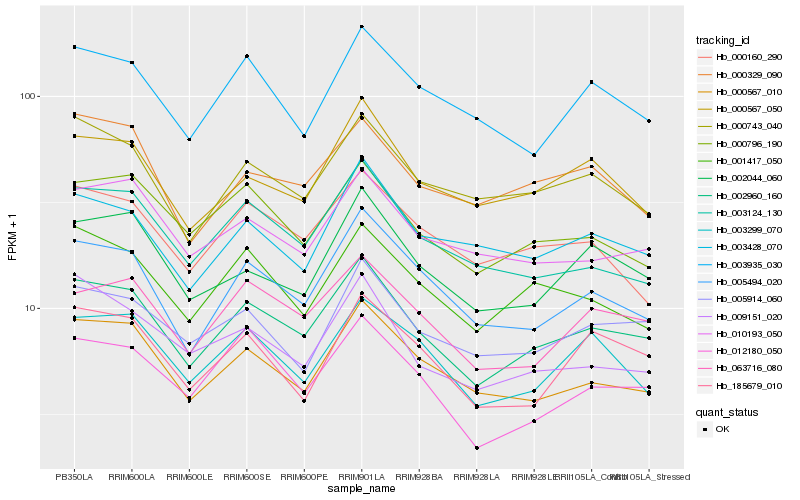
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002960_160 |
0.0 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 35B isoform X2 [Jatropha curcas] |
| 2 |
Hb_005494_020 |
0.0658656385 |
- |
- |
PREDICTED: transmembrane protein 184 homolog DDB_G0279555 [Jatropha curcas] |
| 3 |
Hb_000567_010 |
0.0658940546 |
- |
- |
PREDICTED: uncharacterized protein LOC105631110 [Jatropha curcas] |
| 4 |
Hb_063716_080 |
0.0671896795 |
- |
- |
DNA-directed RNA polymerase III subunit F, putative [Ricinus communis] |
| 5 |
Hb_000796_190 |
0.0727236291 |
- |
- |
hypothetical protein F383_31148 [Gossypium arboreum] |
| 6 |
Hb_000743_040 |
0.0730995484 |
- |
- |
ribosomal RNA methyltransferase, putative [Ricinus communis] |
| 7 |
Hb_002044_060 |
0.0745386499 |
- |
- |
elongation factor ts, putative [Ricinus communis] |
| 8 |
Hb_000567_050 |
0.0749850327 |
- |
- |
PREDICTED: UBP1-associated protein 2C [Jatropha curcas] |
| 9 |
Hb_001417_050 |
0.0752057394 |
- |
- |
PREDICTED: G patch domain-containing protein TGH [Jatropha curcas] |
| 10 |
Hb_005914_060 |
0.0760656979 |
- |
- |
PREDICTED: splicing factor U2af large subunit A [Jatropha curcas] |
| 11 |
Hb_003124_130 |
0.0762865722 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X1 [Jatropha curcas] |
| 12 |
Hb_010193_050 |
0.0781671099 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_012180_050 |
0.0782309645 |
- |
- |
PREDICTED: uncharacterized protein LOC105632355 isoform X3 [Jatropha curcas] |
| 14 |
Hb_003299_070 |
0.0801007318 |
- |
- |
AMP deaminase [Theobroma cacao] |
| 15 |
Hb_003428_070 |
0.0808053142 |
- |
- |
DEAD-box ATP-dependent RNA helicase 46 -like protein [Gossypium arboreum] |
| 16 |
Hb_003935_030 |
0.082211064 |
- |
- |
PREDICTED: 14-3-3-like protein GF14 kappa isoform X2 [Jatropha curcas] |
| 17 |
Hb_009151_020 |
0.0826731462 |
- |
- |
PREDICTED: DNA polymerase delta catalytic subunit [Jatropha curcas] |
| 18 |
Hb_000160_290 |
0.0832698938 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_000329_090 |
0.0837152957 |
- |
- |
arginase, putative [Ricinus communis] |
| 20 |
Hb_185679_010 |
0.0840284876 |
- |
- |
serine/threonine-protein kinase rio2, putative [Ricinus communis] |