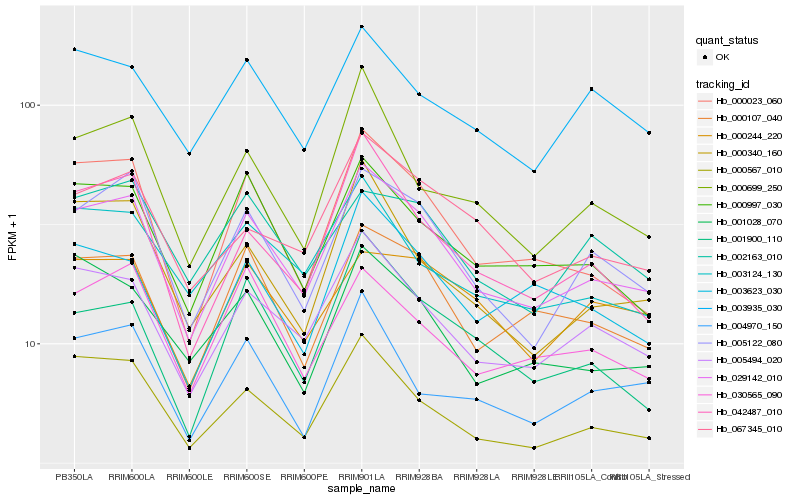
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029142_010 |
0.0 |
- |
- |
PREDICTED: H/ACA ribonucleoprotein complex subunit 4 [Jatropha curcas] |
| 2 |
Hb_005494_020 |
0.0552457912 |
- |
- |
PREDICTED: transmembrane protein 184 homolog DDB_G0279555 [Jatropha curcas] |
| 3 |
Hb_005122_080 |
0.0637566761 |
- |
- |
prokaryotic DNA topoisomerase, putative [Ricinus communis] |
| 4 |
Hb_000567_010 |
0.0673342192 |
- |
- |
PREDICTED: uncharacterized protein LOC105631110 [Jatropha curcas] |
| 5 |
Hb_000107_040 |
0.0678546323 |
- |
- |
nuclear pore complex protein nup153, putative [Ricinus communis] |
| 6 |
Hb_000023_060 |
0.0717832648 |
- |
- |
PREDICTED: uncharacterized protein LOC105633775 [Jatropha curcas] |
| 7 |
Hb_000997_030 |
0.074118047 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 8 |
Hb_042487_010 |
0.0785426841 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 9-like [Jatropha curcas] |
| 9 |
Hb_067345_010 |
0.0823476501 |
- |
- |
PREDICTED: F-box protein CPR30-like [Jatropha curcas] |
| 10 |
Hb_030565_090 |
0.0844241272 |
- |
- |
PREDICTED: DNA repair protein RAD50 [Jatropha curcas] |
| 11 |
Hb_000340_160 |
0.0864154242 |
- |
- |
PREDICTED: elongation factor Tu, mitochondrial [Jatropha curcas] |
| 12 |
Hb_001900_110 |
0.0864321024 |
- |
- |
PREDICTED: replication protein A 70 kDa DNA-binding subunit E [Jatropha curcas] |
| 13 |
Hb_003124_130 |
0.087184499 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001028_070 |
0.0872474879 |
- |
- |
PREDICTED: WD repeat-containing protein 75 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000699_250 |
0.0876684206 |
- |
- |
Poly(rC)-binding protein, putative [Ricinus communis] |
| 16 |
Hb_003623_030 |
0.0876808208 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 14 [Jatropha curcas] |
| 17 |
Hb_003935_030 |
0.0882035612 |
- |
- |
PREDICTED: 14-3-3-like protein GF14 kappa isoform X2 [Jatropha curcas] |
| 18 |
Hb_002163_010 |
0.0895483434 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410 [Jatropha curcas] |
| 19 |
Hb_004970_150 |
0.09018307 |
- |
- |
PREDICTED: uncharacterized protein LOC105633636 [Jatropha curcas] |
| 20 |
Hb_000244_220 |
0.0919047522 |
- |
- |
PREDICTED: bifunctional aspartokinase/homoserine dehydrogenase 1, chloroplastic-like [Jatropha curcas] |