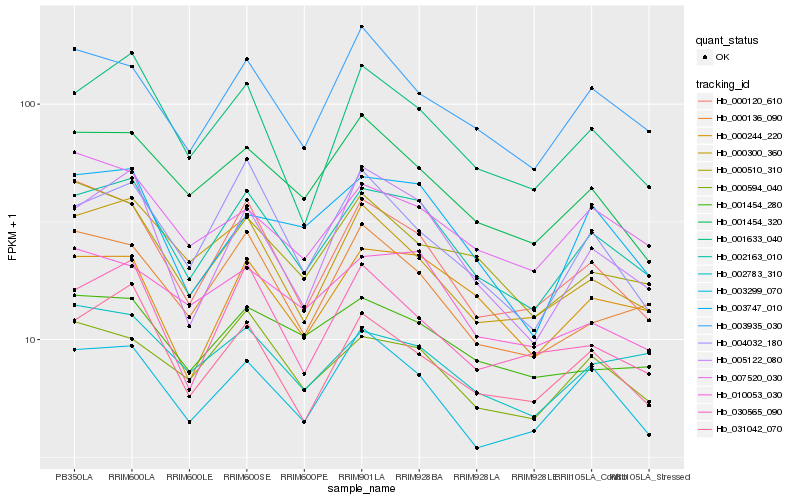
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002163_010 |
0.0 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410 [Jatropha curcas] |
| 2 |
Hb_000594_040 |
0.0702371129 |
- |
- |
hypothetical protein L484_013922 [Morus notabilis] |
| 3 |
Hb_003299_070 |
0.0711923343 |
- |
- |
AMP deaminase [Theobroma cacao] |
| 4 |
Hb_031042_070 |
0.0722441877 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001633_040 |
0.0728597509 |
- |
- |
PREDICTED: protein AUXIN SIGNALING F-BOX 2-like [Jatropha curcas] |
| 6 |
Hb_003935_030 |
0.0745714218 |
- |
- |
PREDICTED: 14-3-3-like protein GF14 kappa isoform X2 [Jatropha curcas] |
| 7 |
Hb_001454_320 |
0.0752534534 |
- |
- |
PREDICTED: protein pleiotropic regulatory locus 1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000244_220 |
0.0753890232 |
- |
- |
PREDICTED: bifunctional aspartokinase/homoserine dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_000120_610 |
0.0781077162 |
- |
- |
mitotic control protein dis3, putative [Ricinus communis] |
| 10 |
Hb_005122_080 |
0.0785046557 |
- |
- |
prokaryotic DNA topoisomerase, putative [Ricinus communis] |
| 11 |
Hb_003747_010 |
0.0804128964 |
- |
- |
hypothetical protein JCGZ_24080 [Jatropha curcas] |
| 12 |
Hb_000136_090 |
0.0806113246 |
- |
- |
programmed cell death protein, putative [Ricinus communis] |
| 13 |
Hb_000300_360 |
0.0819623941 |
- |
- |
PREDICTED: exosome component 10 [Jatropha curcas] |
| 14 |
Hb_010053_030 |
0.0819846087 |
- |
- |
PREDICTED: tRNA (cytosine(34)-C(5))-methyltransferase [Jatropha curcas] |
| 15 |
Hb_002783_310 |
0.0823578505 |
- |
- |
PREDICTED: tRNA (adenine(58)-N(1))-methyltransferase non-catalytic subunit trm6 [Jatropha curcas] |
| 16 |
Hb_001454_280 |
0.0835312052 |
- |
- |
PREDICTED: chaperone protein dnaJ 49 [Jatropha curcas] |
| 17 |
Hb_000510_310 |
0.0839332816 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like [Jatropha curcas] |
| 18 |
Hb_007520_030 |
0.0841387967 |
- |
- |
PREDICTED: peroxisomal membrane protein PEX14 [Jatropha curcas] |
| 19 |
Hb_004032_180 |
0.0848523626 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle [Jatropha curcas] |
| 20 |
Hb_030565_090 |
0.0851807377 |
- |
- |
PREDICTED: DNA repair protein RAD50 [Jatropha curcas] |