| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
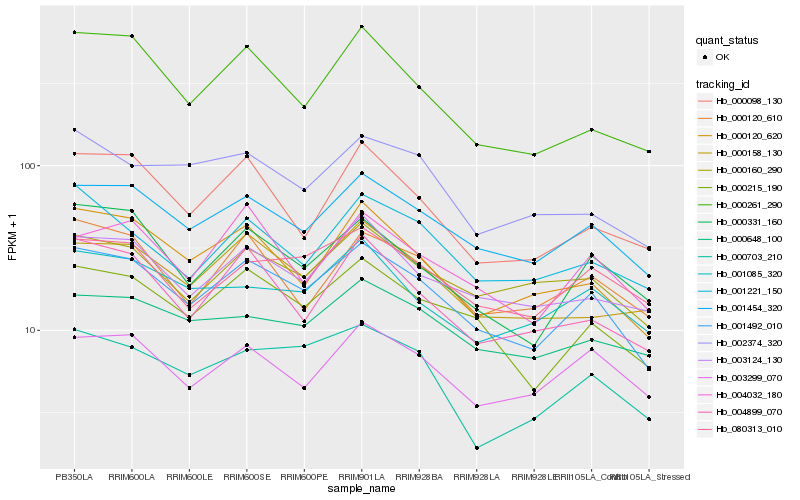
Hb_001492_010 |
0.0 |
- |
- |
PREDICTED: diacylglycerol kinase 3-like isoform X2 [Populus euphratica] |
| 2 |
Hb_001454_320 |
0.0558108235 |
- |
- |
PREDICTED: protein pleiotropic regulatory locus 1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000215_190 |
0.0636094862 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 13-like [Jatropha curcas] |
| 4 |
Hb_000120_620 |
0.0814627963 |
- |
- |
PREDICTED: serine/threonine-protein kinase/endoribonuclease IRE1a isoform X1 [Jatropha curcas] |
| 5 |
Hb_000331_160 |
0.0861780273 |
- |
- |
adenylate kinase 1 chloroplast, putative [Ricinus communis] |
| 6 |
Hb_000160_290 |
0.0884235701 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_003124_130 |
0.0890345612 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000120_610 |
0.0925232362 |
- |
- |
mitotic control protein dis3, putative [Ricinus communis] |
| 9 |
Hb_080313_010 |
0.0934611105 |
- |
- |
PREDICTED: uncharacterized protein LOC105634976 [Jatropha curcas] |
| 10 |
Hb_000261_290 |
0.0942647568 |
- |
- |
eukaryotic translation elongation factor 1-alpha [Hevea brasiliensis] |
| 11 |
Hb_003299_070 |
0.0944469294 |
- |
- |
AMP deaminase [Theobroma cacao] |
| 12 |
Hb_000158_130 |
0.0946411145 |
- |
- |
PREDICTED: uncharacterized protein LOC105647950 isoform X1 [Jatropha curcas] |
| 13 |
Hb_004899_070 |
0.0949674665 |
- |
- |
PREDICTED: RRP12-like protein [Jatropha curcas] |
| 14 |
Hb_004032_180 |
0.0951168741 |
- |
- |
PREDICTED: myosin heavy chain, non-muscle [Jatropha curcas] |
| 15 |
Hb_000098_130 |
0.0968916432 |
- |
- |
PREDICTED: serine/threonine-protein kinase SRK2A [Jatropha curcas] |
| 16 |
Hb_000703_210 |
0.0971756226 |
- |
- |
Inactive protein kinase [Gossypium arboreum] |
| 17 |
Hb_001221_150 |
0.0975919877 |
- |
- |
AMP-activated protein kinase, gamma regulatory subunit, putative [Ricinus communis] |
| 18 |
Hb_000648_100 |
0.0976402252 |
- |
- |
PREDICTED: transcription initiation factor IIA large subunit [Jatropha curcas] |
| 19 |
Hb_001085_320 |
0.0979589888 |
transcription factor |
TF Family: LUG |
WD-repeat protein, putative [Ricinus communis] |
| 20 |
Hb_002374_320 |
0.0987412502 |
- |
- |
hypothetical protein JCGZ_12107 [Jatropha curcas] |