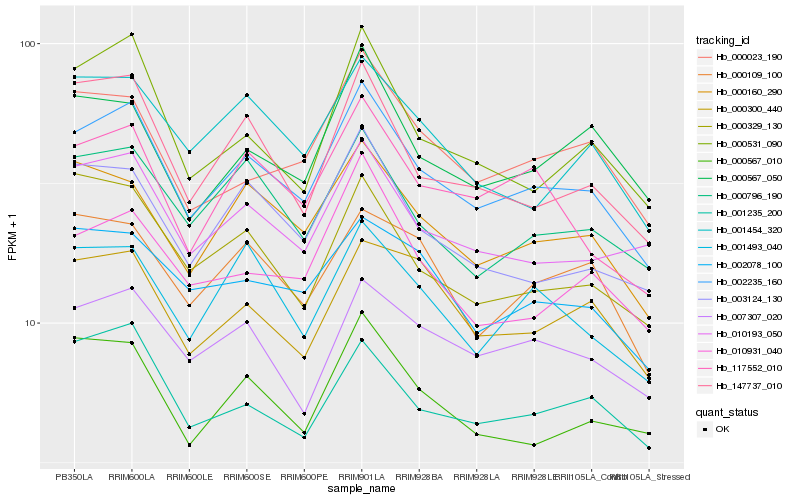
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002235_160 |
0.0 |
- |
- |
PREDICTED: SNF1-related protein kinase catalytic subunit alpha KIN10 [Jatropha curcas] |
| 2 |
Hb_000160_290 |
0.0603676783 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_000796_190 |
0.0689618501 |
- |
- |
hypothetical protein F383_31148 [Gossypium arboreum] |
| 4 |
Hb_117552_010 |
0.069278627 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_001235_200 |
0.0714548274 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003124_130 |
0.0719012539 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X1 [Jatropha curcas] |
| 7 |
Hb_147737_010 |
0.0719929888 |
- |
- |
PREDICTED: arginine/serine-rich coiled-coil protein 2 isoform X2 [Jatropha curcas] |
| 8 |
Hb_001493_040 |
0.0724232942 |
- |
- |
PREDICTED: FACT complex subunit SPT16-like [Jatropha curcas] |
| 9 |
Hb_000567_010 |
0.0725664649 |
- |
- |
PREDICTED: uncharacterized protein LOC105631110 [Jatropha curcas] |
| 10 |
Hb_000023_190 |
0.0727896889 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 11 |
Hb_010193_050 |
0.0728993026 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_007307_020 |
0.0738722753 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_002078_100 |
0.074099987 |
- |
- |
PREDICTED: mannosyl-oligosaccharide glucosidase GCS1-like [Jatropha curcas] |
| 14 |
Hb_000531_090 |
0.0748222687 |
- |
- |
PREDICTED: transcription initiation factor IIF subunit alpha [Jatropha curcas] |
| 15 |
Hb_001454_320 |
0.075658088 |
- |
- |
PREDICTED: protein pleiotropic regulatory locus 1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000567_050 |
0.0759199468 |
- |
- |
PREDICTED: UBP1-associated protein 2C [Jatropha curcas] |
| 17 |
Hb_010931_040 |
0.0765616636 |
- |
- |
PREDICTED: nijmegen breakage syndrome 1 protein [Jatropha curcas] |
| 18 |
Hb_000300_440 |
0.0777692114 |
- |
- |
MIF4G domain and MA3 domain-containing protein isoform 1 [Theobroma cacao] |
| 19 |
Hb_000329_130 |
0.0788515668 |
- |
- |
beta-tubulin cofactor d, putative [Ricinus communis] |
| 20 |
Hb_000109_100 |
0.0789261429 |
- |
- |
PREDICTED: poly [ADP-ribose] polymerase 1 [Jatropha curcas] |