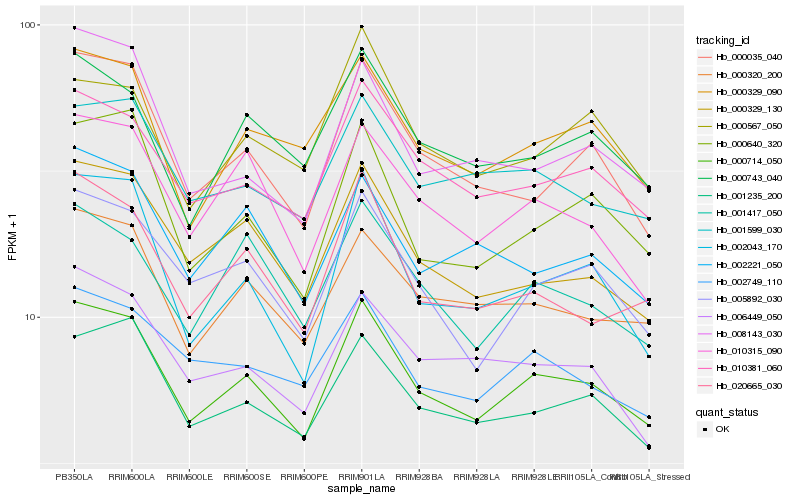
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000714_050 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g16880-like [Jatropha curcas] |
| 2 |
Hb_010381_060 |
0.0531835182 |
- |
- |
DEAD-box ATP-dependent RNA helicase 20 -like protein [Gossypium arboreum] |
| 3 |
Hb_001235_200 |
0.0579465143 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000320_200 |
0.0596162193 |
- |
- |
PREDICTED: U3 small nucleolar RNA-associated protein 6 homolog [Jatropha curcas] |
| 5 |
Hb_005892_030 |
0.0603079025 |
- |
- |
PREDICTED: splicing factor U2af large subunit A [Jatropha curcas] |
| 6 |
Hb_000329_090 |
0.0643060032 |
- |
- |
arginase, putative [Ricinus communis] |
| 7 |
Hb_000329_130 |
0.0649953187 |
- |
- |
beta-tubulin cofactor d, putative [Ricinus communis] |
| 8 |
Hb_000035_040 |
0.0653190636 |
transcription factor |
TF Family: PHD |
PREDICTED: CHD3-type chromatin-remodeling factor PICKLE isoform X1 [Jatropha curcas] |
| 9 |
Hb_000640_320 |
0.0661686503 |
- |
- |
PREDICTED: uncharacterized protein LOC105642644 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000743_040 |
0.0671055507 |
- |
- |
ribosomal RNA methyltransferase, putative [Ricinus communis] |
| 11 |
Hb_001599_030 |
0.0673242129 |
- |
- |
PREDICTED: valine--tRNA ligase [Jatropha curcas] |
| 12 |
Hb_020665_030 |
0.0687209871 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_010315_090 |
0.0690514557 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 14 |
Hb_001417_050 |
0.0699489212 |
- |
- |
PREDICTED: G patch domain-containing protein TGH [Jatropha curcas] |
| 15 |
Hb_002749_110 |
0.0699699155 |
- |
- |
PREDICTED: uncharacterized protein LOC105636001 isoform X1 [Jatropha curcas] |
| 16 |
Hb_006449_050 |
0.0704815859 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ATX2-like [Jatropha curcas] |
| 17 |
Hb_002043_170 |
0.072188191 |
- |
- |
PREDICTED: dentin sialophosphoprotein [Jatropha curcas] |
| 18 |
Hb_002221_050 |
0.0749943461 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP65 isoform X1 [Jatropha curcas] |
| 19 |
Hb_008143_030 |
0.0771776761 |
- |
- |
PREDICTED: uncharacterized protein LOC105650295 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000567_050 |
0.0781311713 |
- |
- |
PREDICTED: UBP1-associated protein 2C [Jatropha curcas] |