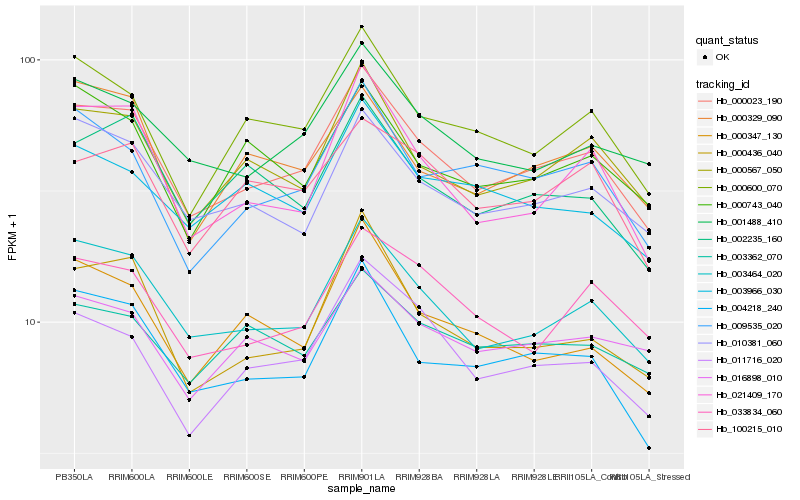
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000023_190 |
0.0 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 2 |
Hb_003464_020 |
0.0433911544 |
- |
- |
srpk, putative [Ricinus communis] |
| 3 |
Hb_000567_050 |
0.0558799028 |
- |
- |
PREDICTED: UBP1-associated protein 2C [Jatropha curcas] |
| 4 |
Hb_004218_240 |
0.0595242294 |
- |
- |
PREDICTED: DNA-directed RNA polymerase V subunit 1 [Jatropha curcas] |
| 5 |
Hb_010381_060 |
0.0612743493 |
- |
- |
DEAD-box ATP-dependent RNA helicase 20 -like protein [Gossypium arboreum] |
| 6 |
Hb_001488_410 |
0.0648561717 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 7 |
Hb_021409_170 |
0.0648723854 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 8 |
Hb_000600_070 |
0.0653219731 |
- |
- |
PREDICTED: leucine aminopeptidase 1-like [Jatropha curcas] |
| 9 |
Hb_000436_040 |
0.0668795164 |
- |
- |
PREDICTED: transportin-1 [Jatropha curcas] |
| 10 |
Hb_009535_020 |
0.069527609 |
- |
- |
PREDICTED: V-type proton ATPase subunit a3-like [Jatropha curcas] |
| 11 |
Hb_011716_020 |
0.0725370171 |
- |
- |
PREDICTED: uncharacterized protein LOC105637783 [Jatropha curcas] |
| 12 |
Hb_002235_160 |
0.0727896889 |
- |
- |
PREDICTED: SNF1-related protein kinase catalytic subunit alpha KIN10 [Jatropha curcas] |
| 13 |
Hb_033834_060 |
0.0728478579 |
- |
- |
DNA-binding bromodomain-containing family protein [Populus trichocarpa] |
| 14 |
Hb_100215_010 |
0.0732768536 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein R isoform X1 [Jatropha curcas] |
| 15 |
Hb_000329_090 |
0.0756251897 |
- |
- |
arginase, putative [Ricinus communis] |
| 16 |
Hb_016898_010 |
0.0774102902 |
- |
- |
PREDICTED: uncharacterized protein PFB0145c [Jatropha curcas] |
| 17 |
Hb_003966_030 |
0.0776142051 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 12-like isoform X7 [Jatropha curcas] |
| 18 |
Hb_000347_130 |
0.0776885619 |
- |
- |
PREDICTED: nuclear pore complex protein NUP107 [Jatropha curcas] |
| 19 |
Hb_003362_070 |
0.0789037658 |
- |
- |
PREDICTED: methyltransferase-like protein 13 [Jatropha curcas] |
| 20 |
Hb_000743_040 |
0.0793031928 |
- |
- |
ribosomal RNA methyltransferase, putative [Ricinus communis] |