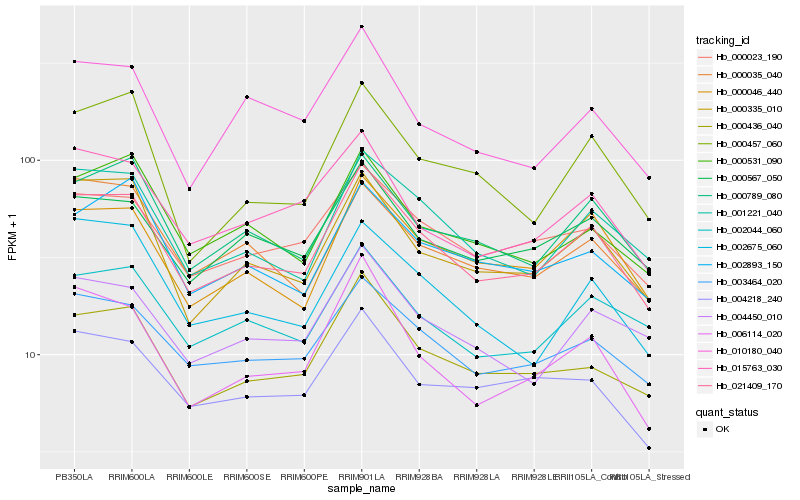
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_021409_170 |
0.0 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 2 |
Hb_003464_020 |
0.0569927209 |
- |
- |
srpk, putative [Ricinus communis] |
| 3 |
Hb_001221_040 |
0.0611856698 |
- |
- |
PREDICTED: nucleolar protein 56-like [Jatropha curcas] |
| 4 |
Hb_000436_040 |
0.0634686755 |
- |
- |
PREDICTED: transportin-1 [Jatropha curcas] |
| 5 |
Hb_000023_190 |
0.0648723854 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 6 |
Hb_000046_440 |
0.0663843013 |
transcription factor |
TF Family: SNF2 |
PREDICTED: putative chromatin-remodeling complex ATPase chain isoform X1 [Jatropha curcas] |
| 7 |
Hb_000789_080 |
0.066786222 |
- |
- |
PREDICTED: arginine--tRNA ligase, cytoplasmic-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000335_010 |
0.0676508541 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 9 |
Hb_000567_050 |
0.0721342561 |
- |
- |
PREDICTED: UBP1-associated protein 2C [Jatropha curcas] |
| 10 |
Hb_006114_020 |
0.0750098249 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_015763_030 |
0.0754870044 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002044_060 |
0.0760051974 |
- |
- |
elongation factor ts, putative [Ricinus communis] |
| 13 |
Hb_000531_090 |
0.0772962549 |
- |
- |
PREDICTED: transcription initiation factor IIF subunit alpha [Jatropha curcas] |
| 14 |
Hb_004218_240 |
0.0775276023 |
- |
- |
PREDICTED: DNA-directed RNA polymerase V subunit 1 [Jatropha curcas] |
| 15 |
Hb_004450_010 |
0.0780625998 |
- |
- |
PREDICTED: methyltransferase-like protein 10 [Jatropha curcas] |
| 16 |
Hb_000035_040 |
0.0807679304 |
transcription factor |
TF Family: PHD |
PREDICTED: CHD3-type chromatin-remodeling factor PICKLE isoform X1 [Jatropha curcas] |
| 17 |
Hb_002893_150 |
0.0811415926 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor ASIL2 [Jatropha curcas] |
| 18 |
Hb_010180_040 |
0.0836916497 |
- |
- |
PREDICTED: prosaposin isoform X1 [Jatropha curcas] |
| 19 |
Hb_002675_060 |
0.0841892105 |
- |
- |
PREDICTED: uncharacterized protein LOC105634954 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000457_060 |
0.0854371829 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |