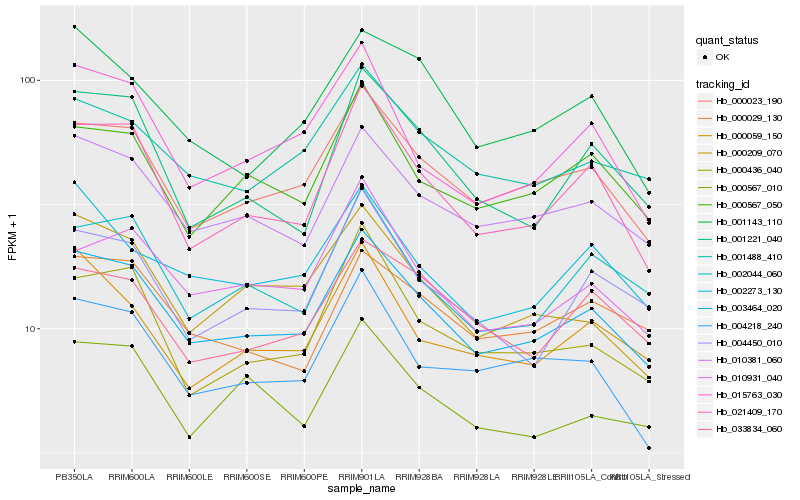
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003464_020 |
0.0 |
- |
- |
srpk, putative [Ricinus communis] |
| 2 |
Hb_000023_190 |
0.0433911544 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 3 |
Hb_010381_060 |
0.0457997423 |
- |
- |
DEAD-box ATP-dependent RNA helicase 20 -like protein [Gossypium arboreum] |
| 4 |
Hb_021409_170 |
0.0569927209 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 5 |
Hb_001488_410 |
0.0612529808 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 6 |
Hb_000567_050 |
0.0641923085 |
- |
- |
PREDICTED: UBP1-associated protein 2C [Jatropha curcas] |
| 7 |
Hb_002044_060 |
0.068935866 |
- |
- |
elongation factor ts, putative [Ricinus communis] |
| 8 |
Hb_004218_240 |
0.0712732231 |
- |
- |
PREDICTED: DNA-directed RNA polymerase V subunit 1 [Jatropha curcas] |
| 9 |
Hb_010931_040 |
0.0716975618 |
- |
- |
PREDICTED: nijmegen breakage syndrome 1 protein [Jatropha curcas] |
| 10 |
Hb_000209_070 |
0.0718840763 |
- |
- |
PREDICTED: chloride channel protein CLC-f isoform X1 [Jatropha curcas] |
| 11 |
Hb_000029_130 |
0.0724949747 |
- |
- |
PREDICTED: ruvB-like protein 1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_002273_130 |
0.0729520934 |
- |
- |
PREDICTED: T-complex protein 1 subunit eta [Gossypium raimondii] |
| 13 |
Hb_004450_010 |
0.073436893 |
- |
- |
PREDICTED: methyltransferase-like protein 10 [Jatropha curcas] |
| 14 |
Hb_015763_030 |
0.0736512717 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000436_040 |
0.0736903179 |
- |
- |
PREDICTED: transportin-1 [Jatropha curcas] |
| 16 |
Hb_000059_150 |
0.0742346082 |
- |
- |
Polynucleotidyl transferase, ribonuclease H-like superfamily protein isoform 2 [Theobroma cacao] |
| 17 |
Hb_033834_060 |
0.0743089999 |
- |
- |
DNA-binding bromodomain-containing family protein [Populus trichocarpa] |
| 18 |
Hb_001221_040 |
0.0754751907 |
- |
- |
PREDICTED: nucleolar protein 56-like [Jatropha curcas] |
| 19 |
Hb_000567_010 |
0.0756983447 |
- |
- |
PREDICTED: uncharacterized protein LOC105631110 [Jatropha curcas] |
| 20 |
Hb_001143_110 |
0.0758117579 |
- |
- |
hypothetical protein JCGZ_12107 [Jatropha curcas] |