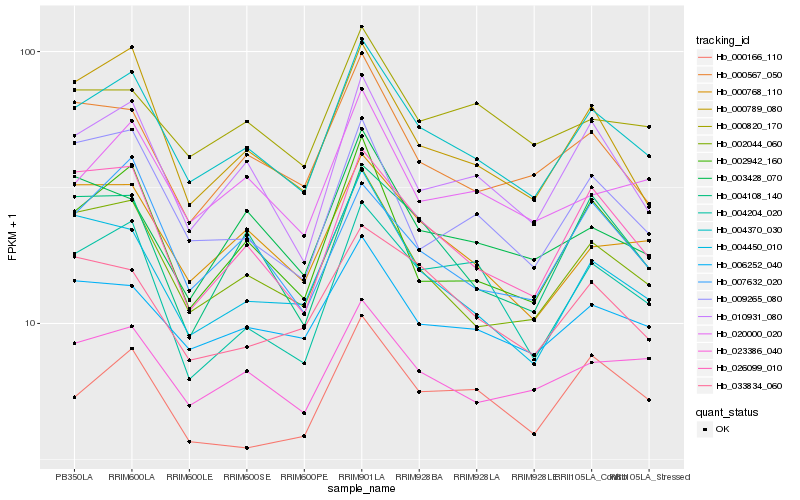
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004370_030 |
0.0 |
- |
- |
PREDICTED: RNA-binding protein 34 [Jatropha curcas] |
| 2 |
Hb_002044_060 |
0.0530241182 |
- |
- |
elongation factor ts, putative [Ricinus communis] |
| 3 |
Hb_010931_080 |
0.0631402696 |
- |
- |
PREDICTED: U4/U6.U5 tri-snRNP-associated protein 2-like [Jatropha curcas] |
| 4 |
Hb_006252_040 |
0.0637414495 |
- |
- |
queuine tRNA-ribosyltransferase, putative [Ricinus communis] |
| 5 |
Hb_026099_010 |
0.0656741537 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF2A isoform X2 [Jatropha curcas] |
| 6 |
Hb_002942_160 |
0.0665597001 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 7 |
Hb_000768_110 |
0.066651726 |
- |
- |
PREDICTED: chromo domain-containing protein LHP1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_023386_040 |
0.0669902614 |
- |
- |
PREDICTED: chaperone protein dnaJ 13-like [Populus euphratica] |
| 9 |
Hb_004450_010 |
0.0705445774 |
- |
- |
PREDICTED: methyltransferase-like protein 10 [Jatropha curcas] |
| 10 |
Hb_000789_080 |
0.071391357 |
- |
- |
PREDICTED: arginine--tRNA ligase, cytoplasmic-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000567_050 |
0.0717952058 |
- |
- |
PREDICTED: UBP1-associated protein 2C [Jatropha curcas] |
| 12 |
Hb_020000_020 |
0.0721414318 |
- |
- |
Troponin C, skeletal muscle, putative [Ricinus communis] |
| 13 |
Hb_007632_020 |
0.0758667848 |
- |
- |
PREDICTED: dnaJ homolog subfamily C member 17 [Jatropha curcas] |
| 14 |
Hb_000166_110 |
0.0766690398 |
transcription factor |
TF Family: SNF2 |
PREDICTED: protein CHROMATIN REMODELING 25 [Jatropha curcas] |
| 15 |
Hb_000820_170 |
0.0783539337 |
- |
- |
PREDICTED: THO complex subunit 4A [Jatropha curcas] |
| 16 |
Hb_004108_140 |
0.0792541756 |
- |
- |
PREDICTED: HUA2-like protein 3 isoform X3 [Jatropha curcas] |
| 17 |
Hb_003428_070 |
0.0793461788 |
- |
- |
DEAD-box ATP-dependent RNA helicase 46 -like protein [Gossypium arboreum] |
| 18 |
Hb_009265_080 |
0.0802364348 |
- |
- |
PREDICTED: apoptosis inhibitor 5 [Jatropha curcas] |
| 19 |
Hb_004204_020 |
0.0806255956 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105640780 [Jatropha curcas] |
| 20 |
Hb_033834_060 |
0.0806488342 |
- |
- |
DNA-binding bromodomain-containing family protein [Populus trichocarpa] |