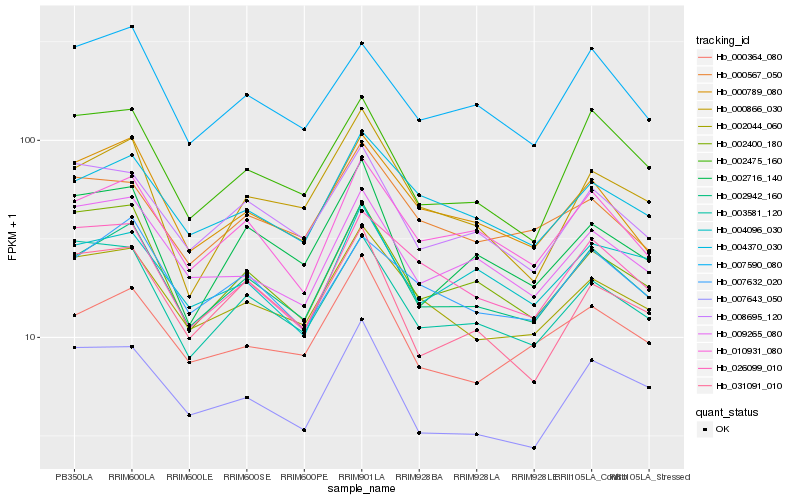
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002942_160 |
0.0 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 2 |
Hb_010931_080 |
0.0558403741 |
- |
- |
PREDICTED: U4/U6.U5 tri-snRNP-associated protein 2-like [Jatropha curcas] |
| 3 |
Hb_002044_060 |
0.0649429453 |
- |
- |
elongation factor ts, putative [Ricinus communis] |
| 4 |
Hb_004370_030 |
0.0665597001 |
- |
- |
PREDICTED: RNA-binding protein 34 [Jatropha curcas] |
| 5 |
Hb_000789_080 |
0.0668084043 |
- |
- |
PREDICTED: arginine--tRNA ligase, cytoplasmic-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000364_080 |
0.0687508385 |
- |
- |
PREDICTED: OTU domain-containing protein 3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_008695_120 |
0.0736699749 |
transcription factor |
TF Family: C2H2 |
PREDICTED: MYST-like histone acetyltransferase 2 [Jatropha curcas] |
| 8 |
Hb_009265_080 |
0.0778814553 |
- |
- |
PREDICTED: apoptosis inhibitor 5 [Jatropha curcas] |
| 9 |
Hb_002400_180 |
0.0778879807 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003581_120 |
0.0783759672 |
- |
- |
PREDICTED: uncharacterized protein LOC105649777 [Jatropha curcas] |
| 11 |
Hb_002475_160 |
0.079135416 |
- |
- |
PREDICTED: 60S ribosomal export protein NMD3 [Jatropha curcas] |
| 12 |
Hb_002716_140 |
0.0799785221 |
- |
- |
PREDICTED: probable VAMP-like protein At1g33475 [Jatropha curcas] |
| 13 |
Hb_007632_020 |
0.0806520105 |
- |
- |
PREDICTED: dnaJ homolog subfamily C member 17 [Jatropha curcas] |
| 14 |
Hb_000866_030 |
0.0813558043 |
- |
- |
hypothetical protein RCOM_1615820 [Ricinus communis] |
| 15 |
Hb_026099_010 |
0.0817701531 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF2A isoform X2 [Jatropha curcas] |
| 16 |
Hb_007590_080 |
0.0821329132 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP19-4 [Malus domestica] |
| 17 |
Hb_031091_010 |
0.0830080331 |
- |
- |
crooked neck protein, putative [Ricinus communis] |
| 18 |
Hb_004096_030 |
0.0832504777 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000567_050 |
0.0836748654 |
- |
- |
PREDICTED: UBP1-associated protein 2C [Jatropha curcas] |
| 20 |
Hb_007643_050 |
0.0837061136 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |