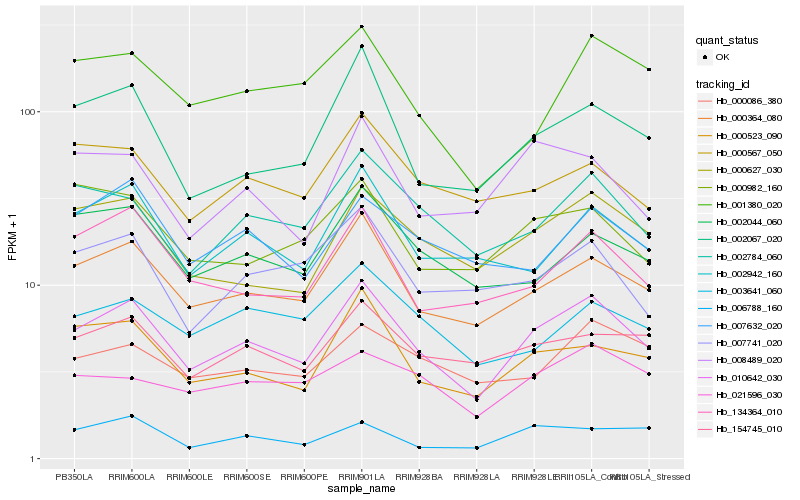
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010642_030 |
0.0 |
- |
- |
zinc phosphodiesterase, putative [Ricinus communis] |
| 2 |
Hb_000364_080 |
0.0827045001 |
- |
- |
PREDICTED: OTU domain-containing protein 3 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002784_060 |
0.1106359443 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA6 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000523_090 |
0.1115029439 |
- |
- |
PREDICTED: protein DJ-1 homolog D-like [Populus euphratica] |
| 5 |
Hb_000627_030 |
0.1115212677 |
- |
- |
amine oxidase, putative [Ricinus communis] |
| 6 |
Hb_002067_020 |
0.1130226629 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002942_160 |
0.1154546067 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 8 |
Hb_134364_010 |
0.1166076177 |
- |
- |
PREDICTED: transcriptional adapter ADA2b isoform X1 [Jatropha curcas] |
| 9 |
Hb_154745_010 |
0.1170594428 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 10 |
Hb_002044_060 |
0.1225233321 |
- |
- |
elongation factor ts, putative [Ricinus communis] |
| 11 |
Hb_001380_020 |
0.1251708818 |
- |
- |
3-isopropylmalate dehydratase, putative [Ricinus communis] |
| 12 |
Hb_000567_050 |
0.1265234041 |
- |
- |
PREDICTED: UBP1-associated protein 2C [Jatropha curcas] |
| 13 |
Hb_021596_030 |
0.1266499376 |
- |
- |
PREDICTED: uncharacterized protein LOC105631841 isoform X1 [Jatropha curcas] |
| 14 |
Hb_008489_020 |
0.1269463181 |
- |
- |
PREDICTED: uncharacterized protein LOC105644663 [Jatropha curcas] |
| 15 |
Hb_000086_380 |
0.1271709745 |
- |
- |
PREDICTED: succinyl-CoA ligase [ADP-forming] subunit beta, mitochondrial [Jatropha curcas] |
| 16 |
Hb_007741_020 |
0.1272001191 |
- |
- |
PREDICTED: uncharacterized protein LOC105643152 [Jatropha curcas] |
| 17 |
Hb_003641_060 |
0.1290416122 |
- |
- |
PREDICTED: uncharacterized protein LOC105638019 [Jatropha curcas] |
| 18 |
Hb_007632_020 |
0.129896915 |
- |
- |
PREDICTED: dnaJ homolog subfamily C member 17 [Jatropha curcas] |
| 19 |
Hb_006788_160 |
0.1312007887 |
- |
- |
hypothetical protein POPTR_0002s04680g [Populus trichocarpa] |
| 20 |
Hb_000982_160 |
0.1321229527 |
- |
- |
PREDICTED: CLIP-associated protein [Jatropha curcas] |