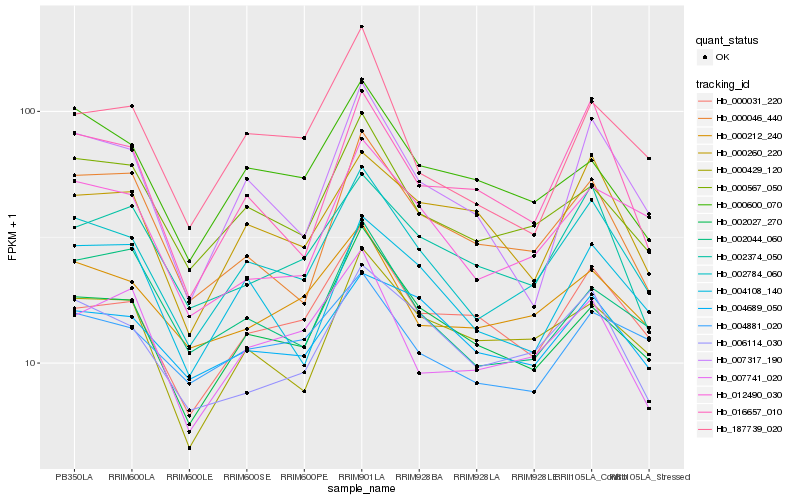
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002784_060 |
0.0 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor TGA6 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000567_050 |
0.0726952302 |
- |
- |
PREDICTED: UBP1-associated protein 2C [Jatropha curcas] |
| 3 |
Hb_002027_270 |
0.076130212 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase BSL1 [Jatropha curcas] |
| 4 |
Hb_006114_030 |
0.0788896041 |
transcription factor |
TF Family: SET |
hypothetical protein CISIN_1g011639mg [Citrus sinensis] |
| 5 |
Hb_012490_030 |
0.0828046069 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000031_220 |
0.085205631 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73B4-like [Jatropha curcas] |
| 7 |
Hb_000212_240 |
0.0856248959 |
- |
- |
short-chain dehydrogenase, putative [Ricinus communis] |
| 8 |
Hb_004108_140 |
0.0870315648 |
- |
- |
PREDICTED: HUA2-like protein 3 isoform X3 [Jatropha curcas] |
| 9 |
Hb_004689_050 |
0.0884465541 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000429_120 |
0.0885901956 |
- |
- |
PREDICTED: DNA-directed RNA polymerase I subunit rpa1 [Jatropha curcas] |
| 11 |
Hb_004881_020 |
0.0889770554 |
- |
- |
PREDICTED: serine/threonine-protein kinase rio2-like [Jatropha curcas] |
| 12 |
Hb_002374_050 |
0.0912373655 |
- |
- |
PREDICTED: auxilin-related protein 1 [Jatropha curcas] |
| 13 |
Hb_000046_440 |
0.0926604903 |
transcription factor |
TF Family: SNF2 |
PREDICTED: putative chromatin-remodeling complex ATPase chain isoform X1 [Jatropha curcas] |
| 14 |
Hb_007741_020 |
0.0938187346 |
- |
- |
PREDICTED: uncharacterized protein LOC105643152 [Jatropha curcas] |
| 15 |
Hb_016657_010 |
0.0939260464 |
- |
- |
PREDICTED: peroxisome biogenesis protein 5 [Jatropha curcas] |
| 16 |
Hb_002044_060 |
0.0939480358 |
- |
- |
elongation factor ts, putative [Ricinus communis] |
| 17 |
Hb_000600_070 |
0.0960846883 |
- |
- |
PREDICTED: leucine aminopeptidase 1-like [Jatropha curcas] |
| 18 |
Hb_007317_190 |
0.0965824493 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI1 [Jatropha curcas] |
| 19 |
Hb_000260_220 |
0.0967154266 |
- |
- |
PREDICTED: RING finger and transmembrane domain-containing protein 2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_187739_020 |
0.0977514747 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |