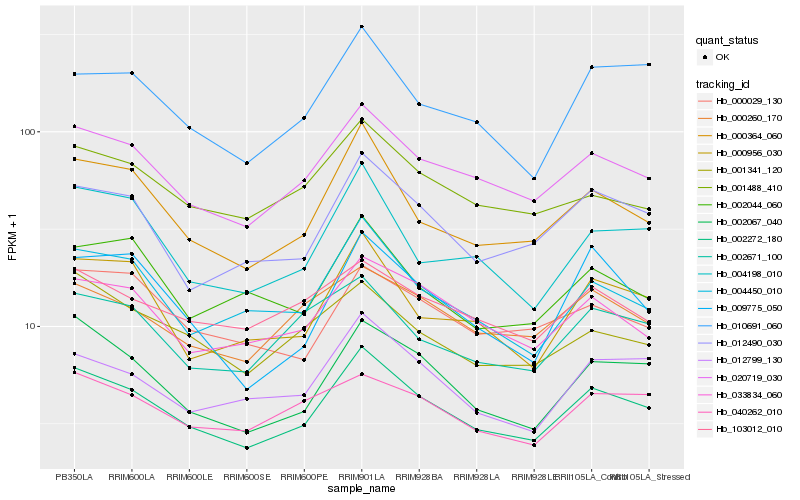
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002272_180 |
0.0 |
- |
- |
hypothetical protein JCGZ_23380 [Jatropha curcas] |
| 2 |
Hb_020719_030 |
0.0499848925 |
- |
- |
PREDICTED: ER membrane protein complex subunit 3-like [Jatropha curcas] |
| 3 |
Hb_000364_060 |
0.0646196472 |
- |
- |
JHL03K20.4 [Jatropha curcas] |
| 4 |
Hb_002067_040 |
0.070637179 |
- |
- |
dihydrodipicolinate reductase, putative [Ricinus communis] |
| 5 |
Hb_012490_030 |
0.0711842862 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004450_010 |
0.0712545112 |
- |
- |
PREDICTED: methyltransferase-like protein 10 [Jatropha curcas] |
| 7 |
Hb_033834_060 |
0.0762513689 |
- |
- |
DNA-binding bromodomain-containing family protein [Populus trichocarpa] |
| 8 |
Hb_012799_130 |
0.0797667582 |
- |
- |
PREDICTED: flap endonuclease GEN-like 2 [Jatropha curcas] |
| 9 |
Hb_000956_030 |
0.0799997742 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 19-like [Jatropha curcas] |
| 10 |
Hb_004198_010 |
0.0814755156 |
- |
- |
Eukaryotic translation initiation factor 2 subunit beta [Theobroma cacao] |
| 11 |
Hb_001488_410 |
0.0832566156 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 12 |
Hb_010691_060 |
0.0858585326 |
- |
- |
PREDICTED: 40S ribosomal protein S6 [Jatropha curcas] |
| 13 |
Hb_000260_170 |
0.0869162218 |
- |
- |
PREDICTED: putative zinc transporter At3g08650 [Cucumis sativus] |
| 14 |
Hb_002671_100 |
0.0869920093 |
- |
- |
PREDICTED: tRNA (guanine-N(7)-)-methyltransferase-like [Jatropha curcas] |
| 15 |
Hb_001341_120 |
0.0882254461 |
- |
- |
hypothetical protein B456_001G130300 [Gossypium raimondii] |
| 16 |
Hb_000029_130 |
0.0900094782 |
- |
- |
PREDICTED: ruvB-like protein 1 isoform X2 [Jatropha curcas] |
| 17 |
Hb_002044_060 |
0.0912260369 |
- |
- |
elongation factor ts, putative [Ricinus communis] |
| 18 |
Hb_009775_050 |
0.0913251634 |
transcription factor |
TF Family: Orphans |
hypothetical protein RCOM_0575190 [Ricinus communis] |
| 19 |
Hb_103012_010 |
0.0914783453 |
- |
- |
PREDICTED: uncharacterized protein LOC105638878 [Jatropha curcas] |
| 20 |
Hb_040262_010 |
0.0918470498 |
- |
- |
PREDICTED: aspartate carbamoyltransferase 1, chloroplastic [Jatropha curcas] |