| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
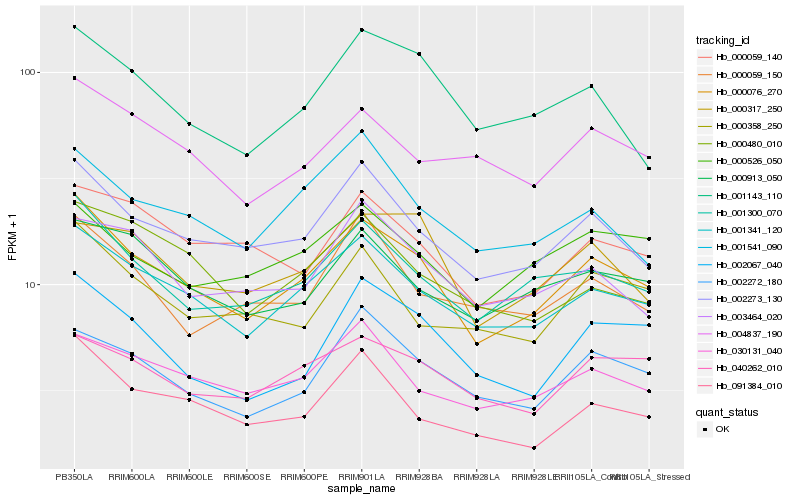
Hb_000076_270 |
0.0 |
- |
- |
PREDICTED: DDRGK domain-containing protein 1 [Populus euphratica] |
| 2 |
Hb_001341_120 |
0.061053152 |
- |
- |
hypothetical protein B456_001G130300 [Gossypium raimondii] |
| 3 |
Hb_002273_130 |
0.0696404464 |
- |
- |
PREDICTED: T-complex protein 1 subunit eta [Gossypium raimondii] |
| 4 |
Hb_001300_070 |
0.0712571891 |
- |
- |
OTU domain-containing protein 6B, putative [Ricinus communis] |
| 5 |
Hb_091384_010 |
0.092038884 |
- |
- |
hypothetical protein JCGZ_09207 [Jatropha curcas] |
| 6 |
Hb_000913_050 |
0.0939010682 |
- |
- |
hypothetical protein JCGZ_17329 [Jatropha curcas] |
| 7 |
Hb_002272_180 |
0.0958102049 |
- |
- |
hypothetical protein JCGZ_23380 [Jatropha curcas] |
| 8 |
Hb_030131_040 |
0.0962895458 |
- |
- |
ribosome biogenesis protein tsr1, putative [Ricinus communis] |
| 9 |
Hb_001143_110 |
0.0971564842 |
- |
- |
hypothetical protein JCGZ_12107 [Jatropha curcas] |
| 10 |
Hb_002067_040 |
0.0976642587 |
- |
- |
dihydrodipicolinate reductase, putative [Ricinus communis] |
| 11 |
Hb_000059_140 |
0.098254856 |
- |
- |
rnase h, putative [Ricinus communis] |
| 12 |
Hb_000358_250 |
0.1002184836 |
- |
- |
PREDICTED: cell division cycle protein 27 homolog B [Jatropha curcas] |
| 13 |
Hb_040262_010 |
0.1003714303 |
- |
- |
PREDICTED: aspartate carbamoyltransferase 1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001541_090 |
0.1012889922 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 15 |
Hb_004837_190 |
0.1017213917 |
- |
- |
PREDICTED: uncharacterized protein LOC105648286 [Jatropha curcas] |
| 16 |
Hb_000526_050 |
0.1022520009 |
- |
- |
PREDICTED: chaperone protein dnaJ 1, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000480_010 |
0.1030146326 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_003464_020 |
0.1037110012 |
- |
- |
srpk, putative [Ricinus communis] |
| 19 |
Hb_000059_150 |
0.1045119641 |
- |
- |
Polynucleotidyl transferase, ribonuclease H-like superfamily protein isoform 2 [Theobroma cacao] |
| 20 |
Hb_000317_250 |
0.1054340893 |
- |
- |
PREDICTED: calcium-dependent protein kinase SK5 isoform X1 [Jatropha curcas] |