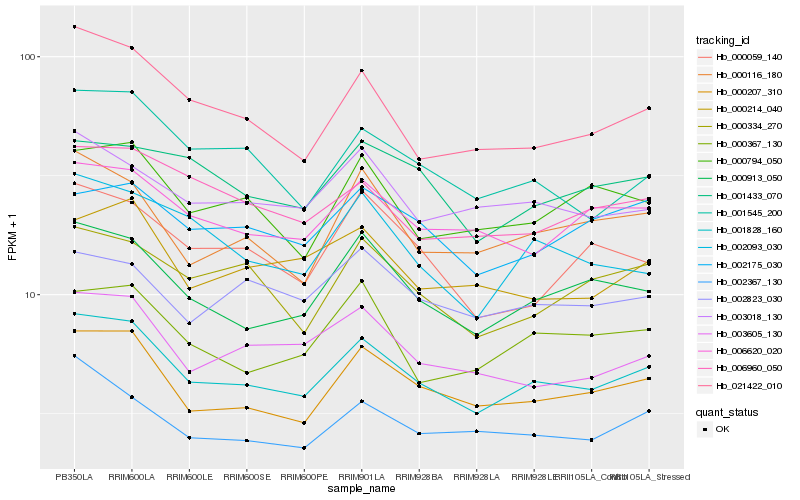
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001828_160 |
0.0 |
- |
- |
hypothetical protein F775_31105 [Aegilops tauschii] |
| 2 |
Hb_000207_310 |
0.0516844446 |
- |
- |
PREDICTED: uncharacterized protein LOC105633932 [Jatropha curcas] |
| 3 |
Hb_000913_050 |
0.0579296548 |
- |
- |
hypothetical protein JCGZ_17329 [Jatropha curcas] |
| 4 |
Hb_021422_010 |
0.0654152024 |
- |
- |
PREDICTED: alanine--glyoxylate aminotransferase 2 homolog 1, mitochondrial [Jatropha curcas] |
| 5 |
Hb_006620_020 |
0.0721414786 |
- |
- |
PREDICTED: uncharacterized protein LOC105638878 [Jatropha curcas] |
| 6 |
Hb_000116_180 |
0.0722657297 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein H isoform X1 [Jatropha curcas] |
| 7 |
Hb_001545_200 |
0.0724464399 |
- |
- |
PREDICTED: uncharacterized oxidoreductase At4g09670 [Jatropha curcas] |
| 8 |
Hb_003605_130 |
0.0727814873 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Populus euphratica] |
| 9 |
Hb_002367_130 |
0.0733264532 |
- |
- |
UNC-50 family protein isoform 1 [Theobroma cacao] |
| 10 |
Hb_003018_130 |
0.0746407228 |
- |
- |
PREDICTED: uncharacterized protein LOC105628777 [Jatropha curcas] |
| 11 |
Hb_006960_050 |
0.076633245 |
- |
- |
PREDICTED: PRKR-interacting protein 1 [Jatropha curcas] |
| 12 |
Hb_002093_030 |
0.0767691261 |
- |
- |
PREDICTED: uncharacterized protein LOC105648523 [Jatropha curcas] |
| 13 |
Hb_000794_050 |
0.0767829535 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 12 [Jatropha curcas] |
| 14 |
Hb_000334_270 |
0.0769183113 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_000214_040 |
0.0778315371 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000367_130 |
0.0778995143 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000059_140 |
0.0821365208 |
- |
- |
rnase h, putative [Ricinus communis] |
| 18 |
Hb_002175_030 |
0.0832173294 |
- |
- |
PREDICTED: snRNA-activating protein complex subunit isoform X2 [Jatropha curcas] |
| 19 |
Hb_001433_070 |
0.0838391607 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1 [Jatropha curcas] |
| 20 |
Hb_002823_030 |
0.0843491384 |
- |
- |
- |