| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
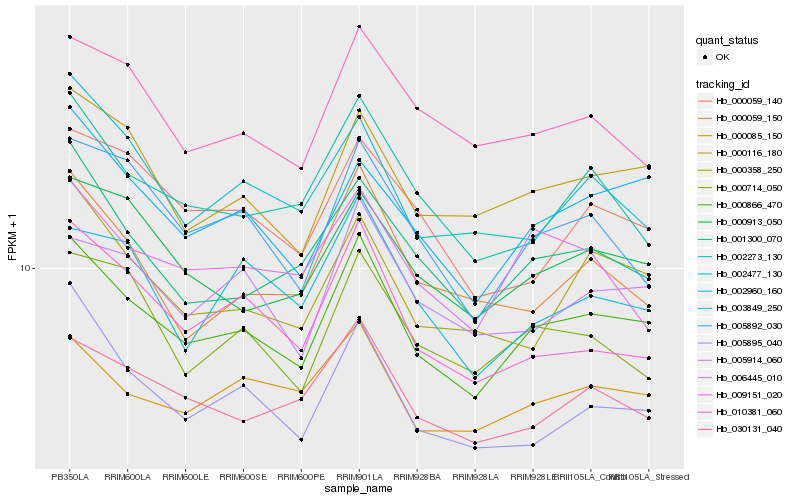
Hb_000866_470 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643015 [Jatropha curcas] |
| 2 |
Hb_000116_180 |
0.0709684179 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein H isoform X1 [Jatropha curcas] |
| 3 |
Hb_003849_250 |
0.0749046055 |
- |
- |
PREDICTED: protein MAK16 homolog [Jatropha curcas] |
| 4 |
Hb_000913_050 |
0.0766004929 |
- |
- |
hypothetical protein JCGZ_17329 [Jatropha curcas] |
| 5 |
Hb_030131_040 |
0.0776201039 |
- |
- |
ribosome biogenesis protein tsr1, putative [Ricinus communis] |
| 6 |
Hb_000085_150 |
0.0786989089 |
- |
- |
PREDICTED: putative BPI/LBP family protein At1g04970 [Jatropha curcas] |
| 7 |
Hb_001300_070 |
0.0801159245 |
- |
- |
OTU domain-containing protein 6B, putative [Ricinus communis] |
| 8 |
Hb_005914_060 |
0.0818961123 |
- |
- |
PREDICTED: splicing factor U2af large subunit A [Jatropha curcas] |
| 9 |
Hb_000358_250 |
0.0830858244 |
- |
- |
PREDICTED: cell division cycle protein 27 homolog B [Jatropha curcas] |
| 10 |
Hb_005892_030 |
0.0833637516 |
- |
- |
PREDICTED: splicing factor U2af large subunit A [Jatropha curcas] |
| 11 |
Hb_002273_130 |
0.0847248006 |
- |
- |
PREDICTED: T-complex protein 1 subunit eta [Gossypium raimondii] |
| 12 |
Hb_009151_020 |
0.0865270959 |
- |
- |
PREDICTED: DNA polymerase delta catalytic subunit [Jatropha curcas] |
| 13 |
Hb_010381_060 |
0.0879476121 |
- |
- |
DEAD-box ATP-dependent RNA helicase 20 -like protein [Gossypium arboreum] |
| 14 |
Hb_000714_050 |
0.0880664817 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g16880-like [Jatropha curcas] |
| 15 |
Hb_000059_140 |
0.0898209884 |
- |
- |
rnase h, putative [Ricinus communis] |
| 16 |
Hb_000059_150 |
0.090031439 |
- |
- |
Polynucleotidyl transferase, ribonuclease H-like superfamily protein isoform 2 [Theobroma cacao] |
| 17 |
Hb_006445_010 |
0.0907681885 |
- |
- |
PREDICTED: uncharacterized protein LOC105640147 [Jatropha curcas] |
| 18 |
Hb_002960_160 |
0.0909490337 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 35B isoform X2 [Jatropha curcas] |
| 19 |
Hb_002477_130 |
0.0910154299 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_005895_040 |
0.0914613757 |
- |
- |
PREDICTED: tRNA threonylcarbamoyladenosine dehydratase [Jatropha curcas] |